Trimethylation Analysis Service
MtoZ Biolabs provides Trimethylation Analysis Service to support accurate identification, quantification, and functional annotation of protein trimethylation events. Using state-of-the-art Orbitrap Fusion Lumos and Q Exactive HF platforms combined with optimized enrichment strategies and advanced bioinformatics interpretation, our service ensures high sensitivity, reproducibility, and reliability. This solution is designed for researchers aiming to investigate the biological significance of trimethylation in diverse cellular contexts.
What is Trimethylation?
Trimethylation is a specific form of lysine or arginine methylation in which three methyl groups are covalently added to a single amino acid residue. Catalyzed by protein methyltransferases, this modification alters protein charge, stability, and binding affinity. Trimethylation of histone lysine residues, such as H3K4me3 and H3K27me3, is a well-established marker of gene activation or repression. Beyond histones, trimethylation occurs on non-histone proteins, influencing transcriptional regulation, chromatin remodeling, signal transduction, and protein-protein interactions. Aberrant trimethylation has been implicated in cancer development, metabolic disorders, and neurological diseases, making its analysis critical for both basic and translational research.
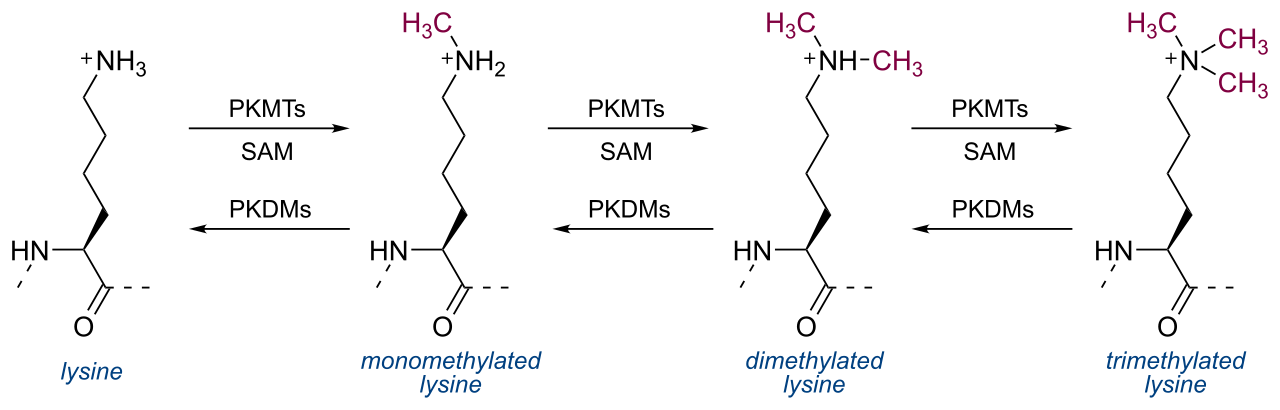
Figure 1. Lysine Trimethylation
Services at MtoZ Biolabs
1. Targeted Protein Trimethylation Analysis
MtoZ Biolabs offers targeted trimethylation analysis for client-designated proteins, providing precise site-level identification and quantitative assessment of trimethylation on specific targets. This service is suited for focused investigations involving defined regulatory proteins, targeted validation studies, or evaluation of modification changes under specific experimental conditions.
2. Trimethylation Proteomics
For broader research needs, MtoZ Biolabs provides trimethylation proteomics to profile trimethylation events across complex biological samples. This proteome-wide approach supports discovery-oriented studies, comparative analyses across biological states or treatments, and the identification of trimethylation patterns relevant to functional regulation or biomarker exploration.
Sample Submission Suggestions
MtoZ Biolabs accepts a wide range of biological materials for trimethylation analysis:
Serum/Plasma: ≥100 µL
Urine: ≥2 mL
Animal Tissue: ≥200 mg
Plant Tissue: ≥10 g
Microorganisms: ≥500 µL
Protein Extracts: ≥5 mg
Cells: ≥1 × 108
Samples should be kept at -80°C and shipped on dry ice. Avoid detergents, salts, or stabilizers that may interfere with LC-MS/MS analysis. If you need further details, our technical support team is happy to assist and provide comprehensive guidance on sample submission.
Why Choose MtoZ Biolabs?
✅ High-Performance Platforms: Equipped with Orbitrap Fusion Lumos and Q Exactive HF for precise trimethylation analysis.
✅ Transparent Pricing: One-time charge with no hidden fees, offering clear cost management.
✅ Robust Data Quality: Strict QC standards and in-depth bioinformatics ensure reliable, reproducible outcomes.
✅ Diverse Sample Compatibility: Validated workflows for tissues, fluids, cell cultures, and purified proteins.
✅ Tailored Project Design: Flexible experimental strategies and customized reports to meet project-specific requirements.
What Could be Included in the Report?
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
Trimethylation plays a central role in regulating transcription, chromatin dynamics, and protein function. Its precise analysis depends on carefully designed enrichment strategies, high-resolution mass spectrometry, and robust bioinformatics interpretation. MtoZ Biolabs provides Trimethylation Analysis Service with an emphasis on data quality and reproducibility, helping researchers obtain reliable insights into complex regulatory networks.
In addition to trimethylation, we also support monomethylation, dimethylation, and a broad spectrum of post-translational modifications including phosphorylation, acetylation, ubiquitination, and glycosylation. This integrated approach provides researchers with dependable tools to explore regulatory mechanisms across diverse biological and disease contexts.
If you are interested in advancing your research with high-quality methylation analysis, we welcome you to contact MtoZ Biolabs to discuss your project needs.
FAQ
Q1: How is trimethylation distinguished from mono- and dimethylation in mass spectrometry?
Each methylation state introduces a distinct incremental mass shift of approximately 14 Da. Monomethylation results in a +14 Da shift, dimethylation adds +28 Da, and trimethylation contributes +42 Da compared to the unmodified residue. High-resolution LC-MS/MS platforms at MtoZ Biolabs can resolve these subtle differences with high mass accuracy and confident site assignment. Optimized fragmentation methods further ensure correct discrimination between different methylation states, reducing false positives and enhancing data reliability.








