4D-Phosphoproteomics Service
Based on a high-resolution mass spectrometry platform (such as LC-MS/MS), MtoZ Biolabs has launched the 4D-phosphoproteomics service which can systematically identify and quantitatively analyze protein phosphorylation sites in complex samples. This service can capture low-abundance or condition-dependent phosphorylation modifications and output high-quality data, including modification site identification results, quantitative difference information, and functional annotations. Through this service, researchers can obtain a more comprehensive and accurate phosphoproteomics map, providing strong data support for signaling pathway analysis, functional studies, and potential biomarker discovery.
Overview
Phosphorylation is a reversible post-translational modification in which a phosphate group is attached to serine, threonine, or tyrosine residues, serving as a key regulator of protein activity, stability, and signaling. Phosphoproteomics systematically investigates phosphorylation sites and their functional roles in cellular processes. 4D-phosphoproteomics, with enhanced resolution and broader coverage, enables precise detection and quantification of low-abundance or condition-dependent phosphorylation events in complex samples. This service has been widely applied in signaling pathway studies, drug target discovery, and biomarker identification, offering reliable data support for understanding protein regulation and advancing biopharmaceutical research.
Analysis Workflow
1. Sample Preparation
Extract and preprocess proteins from samples according to experimental requirements.
2. Protein Digestion and Phosphopeptide Enrichment
Perform enzymatic digestion under optimized conditions and use specific enrichment methods to selectively capture phosphopeptides.
3. 4D Mass Spectrometry Detection
Relying on high-resolution LC-MS/MS combined with 4D separation technology, conduct high-sensitivity identification and quantification of phosphopeptides.
4. Data Analysis
Through database searching and multidimensional data analysis, output site distribution, quantitative differences, and functional annotations to generate a systematic phosphoproteomics map.
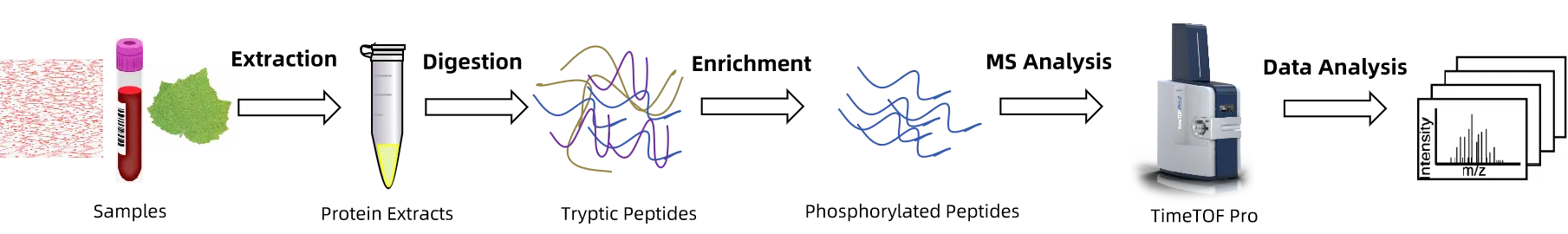
Figure 1. The Workflow of 4D-Phosphoproteomics Analysis.
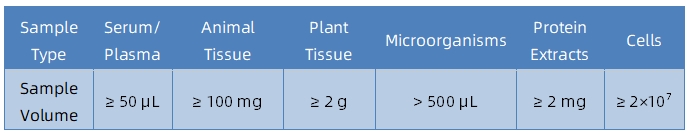
Sample Submission Suggestions
Service Advantages
1. High-Resolution Detection
Relying on advanced high-resolution LC-MS/MS, it can accurately capture low-abundance phosphorylation sites.
2. Comprehensive Coverage
Supports systematic identification and quantification of multiple types of phosphorylation, ensuring comprehensive and reliable results.
3. High Sensitivity and High Reproducibility
Combining multidimensional separation with optimized experimental workflows, it improves detection sensitivity and ensures data consistency.
4. Customized Solutions
Flexibly adjust analysis strategies according to research objectives and sample characteristics to meet different application needs.
Applications
1. Signaling Pathway Research
By systematically analyzing the phosphorylation of key proteins, it can reveal their regulatory roles in cellular signaling networks.
2. Drug Target Screening
The 4D-phosphoproteomics service can be used to evaluate the effects of drug treatment on phosphorylation modifications and identify potential targets.
3. Biomarker Discovery
By screening phosphorylation sites associated with disease states or therapeutic responses, it can support clinical diagnosis and prognosis prediction.
4. Functional Regulation Studies
The 4D-phosphoproteomics service can be used to monitor dynamic phosphorylation events and elucidate their regulatory mechanisms in the cell cycle, metabolism, and stress response.
FAQ
Q1: What Are the Advantages of 4D-Phosphoproteomics Compared with Traditional Phosphoproteomics?
A1: The 4D method integrates ion mobility separation, which effectively improves the detection rate of low-abundance phosphorylation sites in complex samples and significantly enhances the accuracy and reproducibility of quantification.
Q2: Do the Detection Results Include Quantitative Information?
A2: Yes. The service not only provides phosphorylation site identification but also delivers relative or absolute quantitative information. Common methods include Label-free, TMT, iTRAQ, and DIA.
Q3: Will Complex Samples Affect the Detection Performance?
A3: In complex backgrounds, 4D separation can effectively reduce interference, but for highly complex samples, enrichment strategies and multi-step separation are usually recommended to ensure the depth and reliability of the results.








