Protein Backbone Post-translational Modifications Analysis Service
Based on a high-resolution mass spectrometry platform and an advanced liquid chromatography separation system, MtoZ Biolabs has launched the protein backbone post-translational modifications analysis service which enables systematic identification and quantification of various modifications occurring on the protein backbone. This service, combined with specific enrichment and optimized sample processing workflows, can analyze the types of modifications, their site distribution, and abundance differences, and assess their potential impact on the stability of secondary structures and functional performance. The final output data include accurate identification results of modifications, quantitative difference information, and functional annotations, providing researchers with reliable data support.
Overview
Protein backbone post-translational modifications refer to modifications occurring on the chemical groups of the peptide backbone (such as the carbonyl, nitrogen atom of the amide bond, or groups adjacent to the peptide bond). Common forms include deamidation, isomerization, and hydroxylation. These modifications usually alter the local chemical properties of the peptide bond, thereby affecting the hydrogen bond network and spatial orientation of the backbone, which in turn significantly influence the formation and stability of protein secondary structures such as α-helices and β-sheets. Protein backbone post-translational modification analysis is applied in fields such as protein structural biology, aging and stress research, food and nutritional science, and biopharmaceutical quality control, providing reliable data support for studying protein stability, functional realization, and environmental response mechanisms.
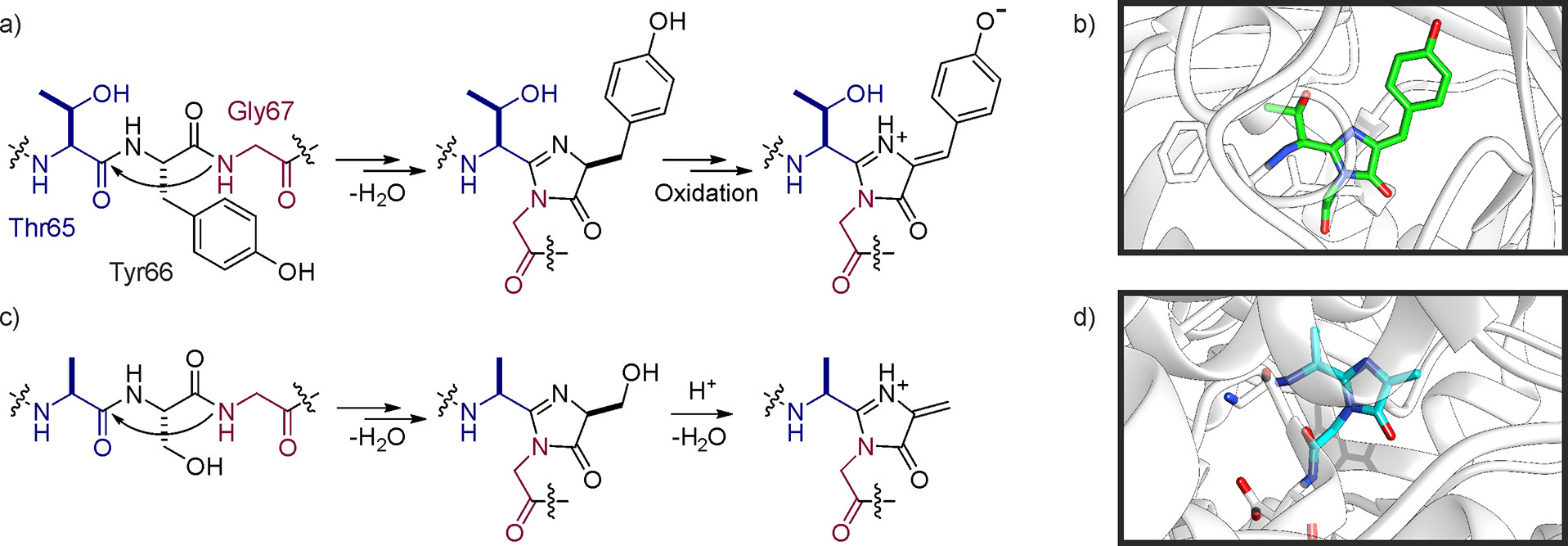
Müller MM. Biochemistry, 2018.
Figure 1. Novel Functions Provided by Backbone Modifications.
Services at MtoZ Biolabs
1. Target Proteins Backbone PTM Analysis
MtoZ Biolabs can perform backbone PTM detection for specific target proteins, identifying the modification types and positions that occur on peptide bonds or backbone atoms. Supported by a high-resolution LC-MS/MS platform, this analysis helps assess how these modifications influence protein folding, stability, and structural characteristics.
2. Proteomics Backbone PTM Analysis
MtoZ Biolabs utilizes high-resolution mass spectrometry combined with multidimensional separation strategies to systematically obtain global backbone PTM types and distribution patterns, and to analyze their structural changes under different conditions. This provides key references for studying structural stability, folding features, and functional regulation.
Analysis Workflow
1. Sample Preparation and Digestion
Samples are pretreated according to their type and subjected to proteolytic digestion.
2. Modified Peptide Enrichment/Labeling
Specific chemical capture or labeling strategies are employed to selectively enrich peptides containing backbone modifications.
3. Mass Spectrometry Detection
High-resolution LC-MS/MS combined with liquid chromatography is used for accurate identification and quantification.
4. Data Analysis
Databases and bioinformatics tools are applied to output modification types, site distribution, abundance differences, and functional annotations.
Service Advantages
1. Comprehensive Coverage
Enables systematic detection of multiple types of backbone modifications, ensuring comprehensive and reliable results.
2. High-Resolution Detection
Based on advanced LC-MS/MS platforms, accurately captures low-abundance backbone modification signals in complex samples.
3. Multi-Method Integration
Combines specific enrichment, labeling, and multiple separation strategies to validate modification sites from different perspectives and enhance result reliability.
4. Customized Solutions
Flexibly adjusts analytical strategies according to experimental goals and sample characteristics to meet diverse research needs.
Applications
1. Protein Stability Research
Protein backbone post-translational modifications analysis service can be used to evaluate protein stability and degradation trends under different conditions.
2. Secondary Structure Impact Analysis
By analyzing backbone modifications, their influence on the formation and stability of secondary structures such as α-helices or β-sheets can be assessed.
3. Biomarker Discovery
Protein backbone post-translational modifications analysis service can be applied to identify potential biomarkers associated with drug action or disease states.
4. Functional Research
By studying the role of backbone modifications in signal pathway regulation and cellular functions, data support can be provided for target research.
FAQ
Q1: How to Distinguish Backbone Modifications from Side-Chain Modifications?
A1: Backbone modifications mainly occur at the carbonyl or amino sites of the peptide bond, directly affecting the chemical properties of the peptide bond, while side-chain modifications act on specific amino acid residues. High-resolution mass spectrometry combined with specific fragmentation modes (such as ETD and EThcD) can differentiate between the two and confirm modification types through database annotation.
Q2: What Are the Differences in Detecting Backbone Modifications Across Different Sample Types?
A2: Complex samples have significant background interference and require stronger separation and enrichment strategies, whereas purified protein samples are more suitable for high-precision site identification and abundance quantification of modifications.








