DIA/SWATH-Based Quantitative PTM Service
Post-translational modifications (PTMs) are essential regulatory mechanisms that influence nearly every aspect of protein function, including activity, stability, localization, and molecular interactions. Quantitative analysis of PTMs provides critical insights into signaling networks, disease mechanisms, and therapeutic responses.
MtoZ Biolabs offers high-performance DIA/SWATH-Based Quantitative PTM Service that enables comprehensive, label-free quantification of modified peptides with superior accuracy and reproducibility. By integrating Data-Independent Acquisition (DIA) and Sequential Window Acquisition of All Theoretical Mass Spectra (SWATH) workflows on cutting-edge LC–MS/MS systems, we capture complete fragment ion information for every detectable peptide, ensuring high coverage, minimal missing values, and excellent quantitative consistency across biological samples.
Overview
Data-Independent Acquisition (DIA) and Sequential Window Acquisition of All Theoretical Mass Spectra (SWATH) are advanced proteomic strategies designed to capture all peptide fragment ions within predefined mass windows. Unlike data-dependent approaches (DDA) that rely on selective precursor targeting, DIA/SWATH systematically collects MS/MS spectra for every detectable peptide, producing a complete and reproducible digital record of the proteome.
This acquisition model provides exceptional quantitative accuracy, reproducibility, and sensitivity across diverse biological samples and experimental conditions. When paired with high-resolution LC–MS/MS instrumentation and curated spectral libraries, DIA/SWATH enables precise and reproducible quantification of site-specific modifications such as phosphorylation, acetylation, ubiquitination, methylation, glycosylation, and succinylation.
The method complements discovery-based DDA workflows by offering deeper coverage, stronger quantification consistency, and scalability for large-cohort or longitudinal studies.
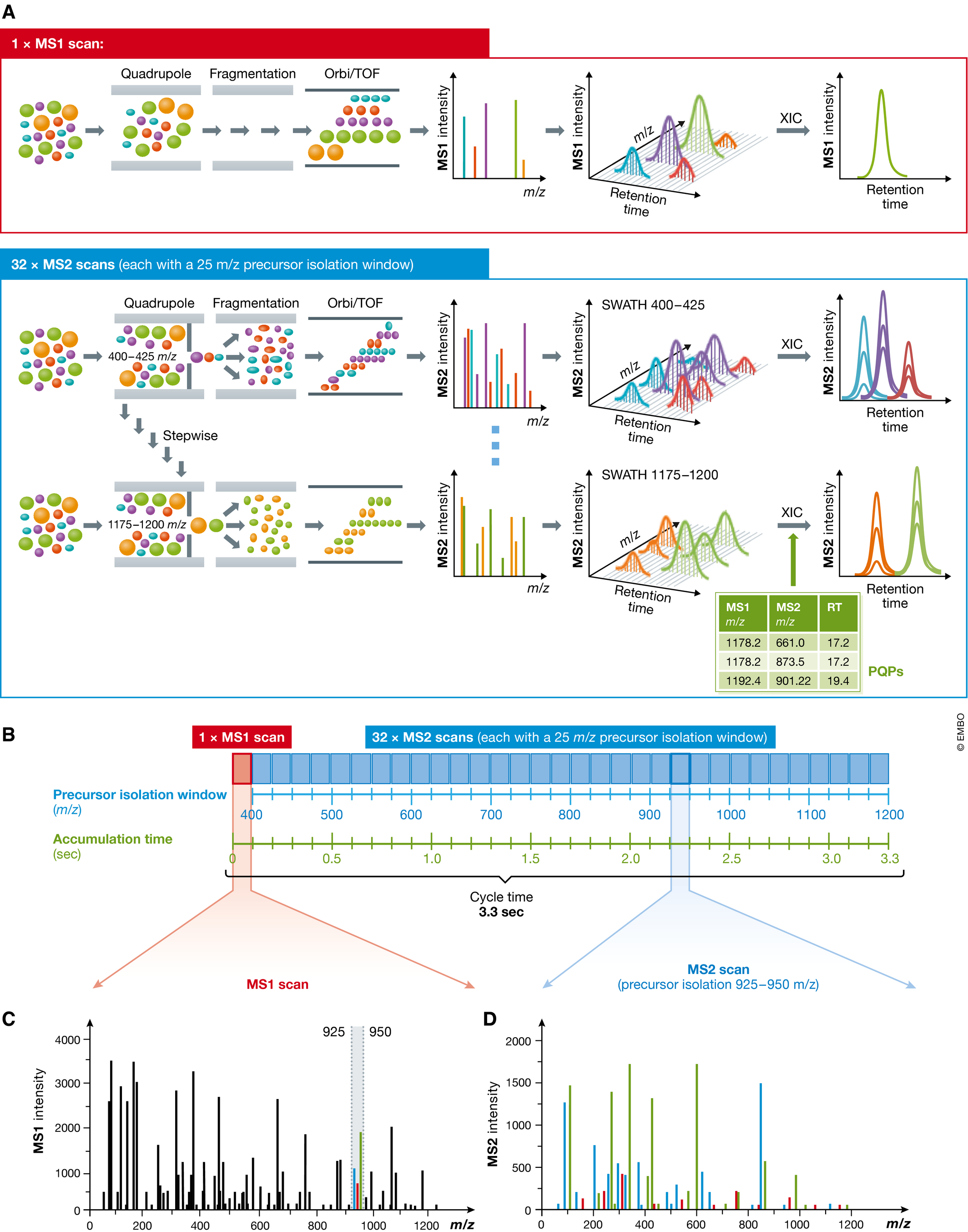
Ludwig, C. et al. Mol Syst Biol. 2018.
Figure 1. Principle of Sequentially Windowed Data-Independent Acquisition in SWATH-MS
Analysis Workflow
MtoZ Biolabs combines optimized enrichment chemistries, high-resolution LC–MS/MS platforms, and advanced bioinformatics pipelines to deliver accurate, reproducible DIA/SWATH-Based Quantitative PTM Service.
1. Experimental Consultation
Customized project design addressing research objectives, sample type, and targeted modification categories.
2. Sample Preparation and PTM Enrichment
Proteins are digested and enriched using modification-specific methods such as IMAC or TiO₂ for phosphopeptides, anti-acetyl-lysine and K-ε-GG immunoaffinity for acetylation and ubiquitination, and HILIC or lectin-based approaches for glycosylation.
3. DIA/SWATH LC–MS/MS Acquisition
High-resolution Orbitrap Exploris or timsTOF platforms acquire comprehensive, unbiased fragment ion spectra, ensuring consistent quantification across replicates.
4. Data Processing and Quantification
Spectral libraries generated from DDA or direct-DIA runs are processed through Spectronaut or DIA-NN for precise peak extraction, normalization, and statistical comparison.
5. Bioinformatics Interpretation
Identified PTM sites are annotated through Gene Ontology and KEGG enrichment, and visualized with volcano plots, heat maps, and pathway diagrams.
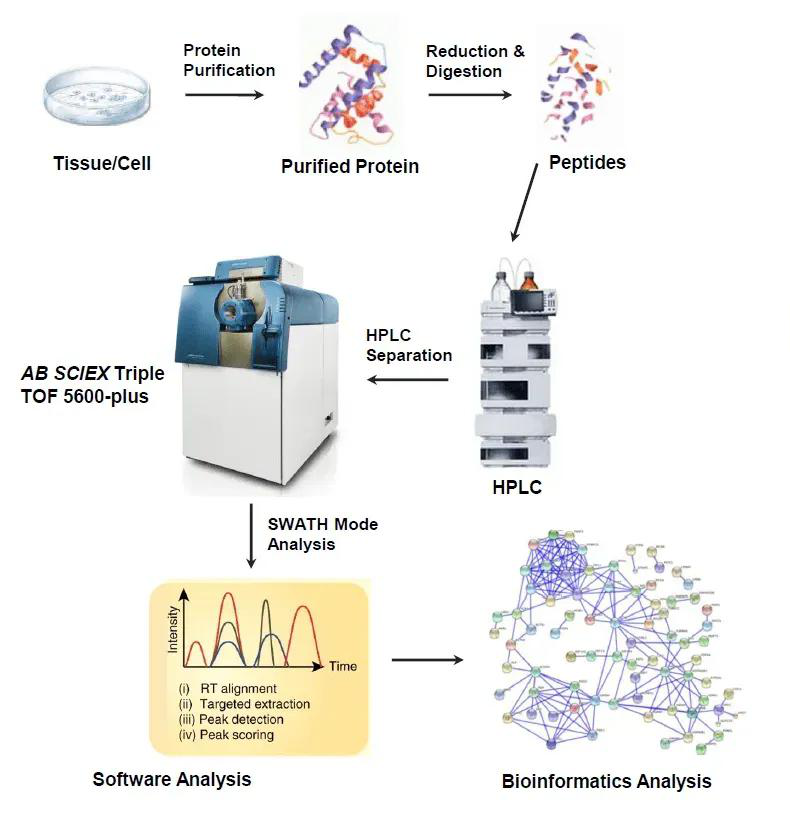
Figure 1. DIA/SWATH-Based Quantitative Analysis Workflow
Why Choose MtoZ Biolabs?
✔ Comprehensive Quantification: DIA/SWATH ensures unbiased, full-spectrum peptide coverage.
✔ High Reproducibility: Uniform acquisition reduces variability and missing data.
✔ Enhanced Sensitivity: Detects low-abundance PTMs with high precision.
✔ Integrated Informatics: Advanced data processing and biological annotation pipelines.
✔ Flexible Scalability: Suitable for discovery research, validation, and large-sample cohort studies.
Sample Submission Suggestions
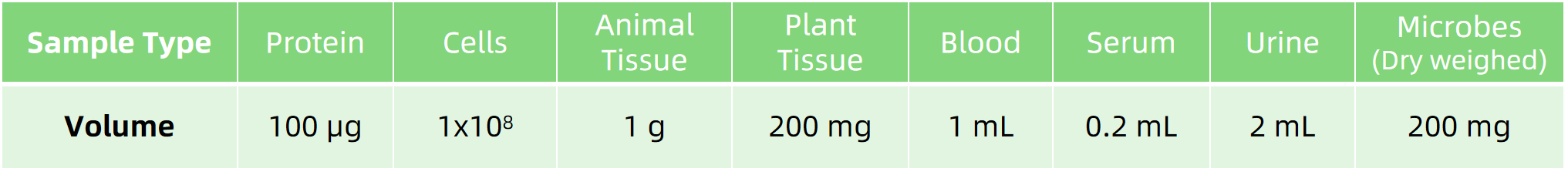
Note: Provide details on sample collection and handling. If you need further details, our technical support team is happy to assist and provide comprehensive guidance on sample submission.
Applications
● Comprehensive PTM Profiling: Quantify phosphorylation, acetylation, ubiquitination, and other modifications across complex proteomes.
● Signaling Pathway Mapping: Characterize site-specific modification dynamics and identify key regulatory enzymes or complexes.
● Drug Mechanism and Target Validation: Evaluate PTM alterations induced by candidate therapeutics or environmental stimuli.
● Comparative Proteomics and Biomarker Discovery: Identify PTM biomarkers distinguishing disease and control samples.
● Quantitative Systems Biology: Integrate PTM data with multi-omics datasets to model cellular signaling and molecular regulation.
Data-independent acquisition has revolutionized quantitative proteomics by combining comprehensive peptide detection with unmatched reproducibility. Leveraging this technology, MtoZ Biolabs delivers a robust DIA/SWATH-Based Quantitative PTM Service that enables site-specific quantification and large-scale comparative analysis with exceptional accuracy. Our integrated workflow helps researchers uncover PTM-mediated regulation, identify therapeutic targets, and drive discoveries in molecular and translational proteomics.
For more information or project consultation, contact MtoZ Biolabs today. Our scientific specialists are ready to assist you.
What Could be Included in the Report?
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files








