Phosphorylation Site Identification Service
Accurate identification of phosphorylation sites is essential for understanding how proteins are regulated, how signaling pathways function, and how cellular processes respond to external stimuli. Phosphorylation Site Identification Service at MtoZ Biolabs helps researchers pinpoint the exact residues modified, uncover kinase–substrate relationships, and generate high-value data for disease research, drug discovery, and biomarker development.
Overview
Phosphorylation is one of the most widespread post-translational modifications in eukaryotes. Protein kinases add negatively charged phosphate groups to serine (Ser), threonine (Thr), or tyrosine (Tyr) residues, while phosphatases remove them in reversible cycles. These modifications influence protein conformation, activity, localization, and interactions, thereby regulating gene expression, intracellular signal transduction, cell proliferation, differentiation, apoptosis, and more. Given the complexity and dynamic nature of phosphorylation, precise mapping of phosphorylation sites provides insights into regulatory mechanisms at the molecular level.
Services at MtoZ Biolabs
1. Targeted Phosphorylation Site Analysis
MtoZ Biolabs provides targeted phosphorylation site analysis for client-defined proteins, offering focused characterization of specific phosphorylation events. This service supports precise site confirmation, targeted quantification, and evaluation of phosphorylation changes associated with defined treatments or biological conditions. It is well suited for mechanism-driven studies, validation of key regulatory proteins, and projects requiring high specificity for individual targets or a small protein panel.
2. Phosphorylation Site Proteomics
MtoZ Biolabs offers phosphorylation site proteomics for global, system-wide assessment of phosphorylation across complex biological samples. This service enables large-scale mapping and quantitative comparison of phosphorylation patterns across tissues, cell populations, or experimental groups. It is designed for discovery research, pathway-level analysis, and identification of phosphorylation signatures with potential functional or translational relevance in disease and therapeutic studies.

Figure 1. Phosphorylation Site Identification Workflow by Mass Spectrometry
MtoZ Biolabs performs these analyses using high-resolution Orbitrap platforms including Q Exactive HF, Fusion, and Fusion Lumos together with phosphopeptide enrichment strategies such as TiO₂, IMAC, SCX, and immunoprecipitation to ensure confident site detection and localization. Computational assessment is available as an additional option to support candidate site prioritization and complement experimental identification.
Why Choose MtoZ Biolabs?
✔ Advanced MS Platforms: Orbitrap Fusion, Lumos, and Q Exactive HF combined with Nano-LC deliver superior sensitivity and resolution.
✔ High-Confidence Site Localization: State-of-the-art algorithms minimize ambiguity in site assignment.
✔ Integrated Workflow: From enrichment to site mapping and bioinformatics, ensuring reproducibility and reliability.
✔ Flexible Project Design: Customized strategies tailored to research focus and sample type.
✔ Transparent Pricing: One-time charge with no hidden costs.
Sample Submission Suggestions
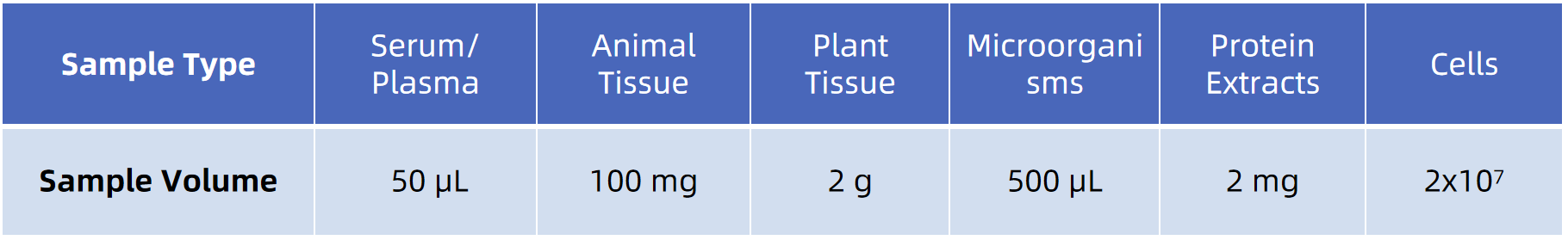
Note: Provide details on sample collection and handling. If you need further details, our technical support team is happy to assist and provide comprehensive guidance on sample submission.
Applications
The Phosphorylation Site Identification Service supports a wide range of research needs:
1. Signal Transduction Research: Mapping phosphorylation events in pathways such as MAPK and PI3K-Akt.
2. Disease Mechanism Studies: Linking site-specific phosphorylation changes to cancer, neurodegenerative disorders, and metabolic diseases.
3. Drug Development: Identifying kinase substrates and monitoring phosphorylation-dependent drug responses.
4. Biomarker Discovery: Detecting phosphorylation signatures with diagnostic or prognostic potential.
5. Plant and Agricultural Research: Investigating phosphorylation in stress adaptation and crop improvement.
FAQ
Q1: Can Both Monophosphorylation and Polyphosphorylation Be Quantified?
Yes. Our layered detection strategy distinguishes between monophosphorylated and polyphosphorylated peptides, minimizing ionization bias and providing more accurate quantitative data.
Q2: Can Phosphorylation of Serine, Threonine, and Tyrosine be Identified Simultaneously?
Yes. With high-resolution LC-MS/MS and optimized fragmentation strategies, MtoZ Biolabs can precisely define the phosphorylation of Ser, Thr, and Tyr residues with high confidence scores.
What Could be Included in the Report?
1. Comprehensive experimental details
2. Materials, instruments, and methods
3. High-quality spectra and visual charts
4. Site-specific phosphorylation identification with detailed annotations
5. Bioinformatics analysis
6. Raw data files








