Ubiquitination Site Identification Service
Precise identification of ubiquitination sites is crucial for understanding how proteins are regulated, targeted for degradation, or functionally modified in different cellular contexts. Ubiquitination Site Identification Service at MtoZ Biolabs provides researchers with accurate residue-level mapping of ubiquitination, enabling detailed insights into protein turnover, signal transduction, immune regulation, and disease mechanisms.
Overview
Ubiquitination is one of the most versatile post-translational modifications (PTMs), in which ubiquitin, a 76-amino acid polypeptide, is covalently attached to lysine residues of substrate proteins. This process is catalyzed sequentially by E1 activating enzymes, E2 conjugating enzymes, and E3 ligases, while deubiquitinating enzymes (DUBs) reverse the modification.
Ubiquitination regulates numerous cellular processes, including proteasomal degradation, DNA repair, vesicle trafficking, autophagy, and immune signaling. Dysregulation of ubiquitination pathways has been linked to cancer, neurodegeneration, metabolic disorders, and infectious diseases. Given the diversity of ubiquitin chain types (mono- and poly-ubiquitination) and linkage patterns (e.g., K48, K63, K11, linear), accurate mapping of ubiquitination sites is essential for deciphering functional outcomes and guiding therapeutic research.
Services at MtoZ Biolabs
1. Targeted Ubiquitination Site Analysis
MtoZ Biolabs provides targeted analysis of ubiquitination sites on client-defined proteins. This service offers precise site-level identification and quantitative assessment for selected targets and is suitable for focused studies that require verification of specific ubiquitination events or evaluation of modification changes under defined biological conditions.
2. Ubiquitination Site Proteomics
MtoZ Biolabs offers proteome-wide ubiquitination site profiling for complex biological samples. This service enables large-scale mapping and comparative analysis of ubiquitination patterns across tissues, cell types, or treatment groups, supporting discovery-driven research and pathway-level investigations.
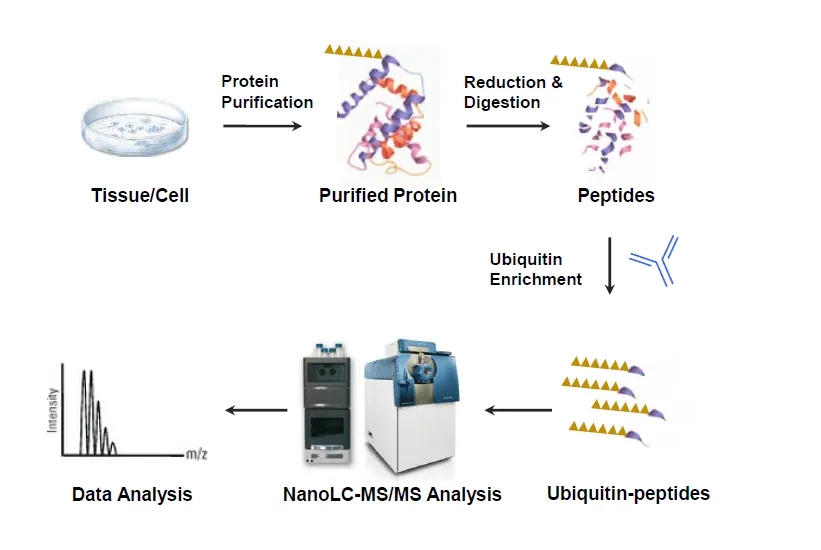
Figure 1. Ubiquitination Site Identification Workflow by Mass Spectrometry
MtoZ Biolabs supports these services with high resolution Orbitrap platforms and K ε GG remnant motif based enrichment to ensure reliable detection of ubiquitinated peptides. Computational assessments are available to assist with motif evaluation, linkage type analysis, and interpretation of results in the context of broader regulatory networks.
Why Choose MtoZ Biolabs?
✔ Advanced MS Platforms: Orbitrap Fusion, Lumos, and Q Exactive HF with Nano-LC ensure ultra-high sensitivity and resolution.
✔ Specific Enrichment: K-ε-GG antibody-based enrichment captures ubiquitinated peptides with high affinity and specificity.
✔ Comprehensive Data Analysis: Bioinformatics tools provide motif analysis, pathway enrichment, and network interpretation.
✔ Flexible Project Design: Customized solutions tailored to different biological systems and research needs.
✔ Transparent Pricing: One-time charge with no hidden costs.
Sample Submission Suggestions
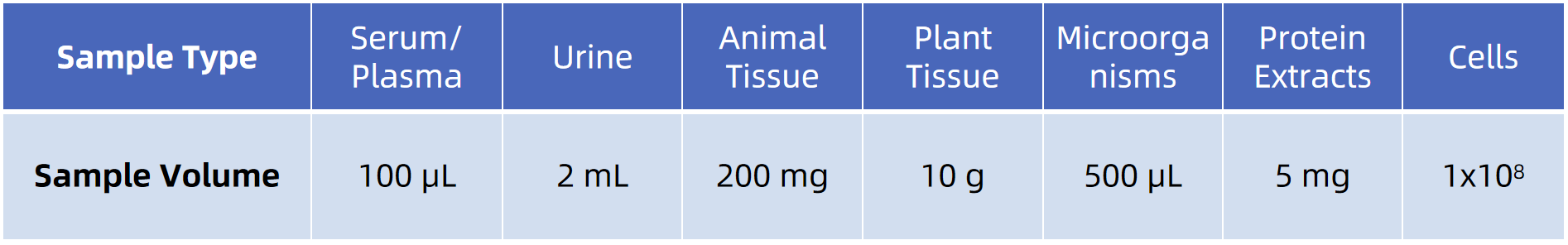
Note: Provide details on sample collection and handling. If you need further details, our technical support team is happy to assist and provide comprehensive guidance on sample submission.
Applications
Our Ubiquitination Site Identification Service supports a wide range of biological and translational research areas:
Protein Degradation and Turnover: Defining ubiquitination sites involved in proteasomal targeting and stability control.
Signal Transduction: Revealing ubiquitin-dependent regulation of kinase cascades and immune signaling pathways.
Disease Research: Mapping dysregulated ubiquitination sites in cancer, neurodegeneration, and metabolic disorders to uncover therapeutic targets.
Drug Discovery: Identifying E3 ligase-substrate relationships for targeted protein degradation strategies, such as PROTACs and molecular glues.
What Could be Included in the Report?
1. Experimental Design & Sample Metadata
2. Enrichment Strategy, Antibodies & Methods
3. LC–MS/MS Acquisition & Quality Control
4. Identification & Site Localization
5. Bioinformatics Interpretation
6. Deliverables & Raw Data
Ubiquitination Site Identification Service at MtoZ Biolabs empowers researchers to uncover the regulatory roles of ubiquitination with precision and confidence. Our integration of enrichment, Orbitrap-based proteomics, and advanced bioinformatics delivers high-value datasets that accelerate discovery from molecular mechanisms to therapeutic applications.
Beyond ubiquitination, MtoZ Biolabs provides a full portfolio of PTM-omics services, supporting comprehensive studies of protein regulation. Contact us today to learn how our Ubiquitination Site Identification Service can advance your research.








