4D-Methylation Proteomics Service
Based on a high-resolution mass spectrometry platform combined with ion mobility separation technology, MtoZ Biolabs has launched the 4D-methylation proteomics service which enables systematic detection and quantitative analysis of protein methylation modifications in complex biological samples. This service, through specific enrichment strategies and optimized experimental workflows, can accurately identify different types of methylation sites and capture their abundance changes. The final output data include modification site identification results, quantitative difference analysis, and functional annotation, helping researchers comprehensively map methylation modification profiles and providing reliable data support for epigenetics research, signaling pathway analysis, and cellular function exploration.
Overview
Methylation is an important form of post-translational modification, commonly occurring on lysine or arginine residues, and regulates protein spatial conformation, complex assembly, and interactions with nucleic acids or other proteins through mono-, di-, or tri-methylation. Methylation proteomics analysis can achieve accurate detection and quantification of low-abundance methylation sites in complex backgrounds, comprehensively depict modification features and dynamic changes, and generate systematic modification maps. It has been widely applied in epigenetics, cellular functional mechanism analysis, signaling network research, and potential biomarker discovery, providing high-quality data support for scientific research and pharmaceutical development.
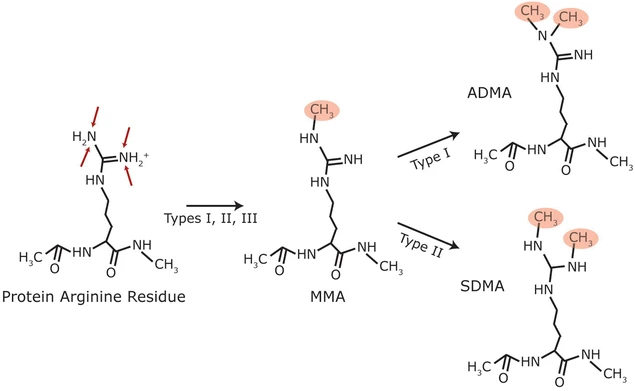
Raposo, A E. et al. Cell Division, 2018.
Figure 1. Types of Mammalian Protein Arginine Methylation.
Analysis Workflow
1. Sample Preparation
Proteins are extracted and pretreated from samples according to experimental requirements.
2. Protein Digestion and Peptide Enrichment
Proteins are enzymatically digested under optimized conditions, and methylated peptides are selectively enriched using specific strategies.
3. 4D Mass Spectrometry Detection
High-resolution LC-MS/MS combined with ion mobility separation technology is used to achieve highly sensitive identification and quantitative analysis.
4. Data Analysis
Databases and bioinformatics tools are applied to output results including methylation site distribution, quantitative differences, and functional annotation.
Sample Submission Suggestions
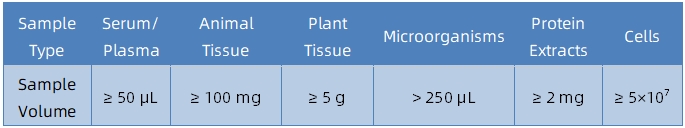
Service Advantages
1. High Resolution
Utilizes advanced mass spectrometry and ion mobility separation to achieve accurate detection of low-abundance methylation sites.
2. Comprehensive Modification Analysis
Covers multiple types of methylation and provides systematic identification and quantitative results.
3. Sensitivity and Stability
Combines enrichment strategies and optimized workflows to ensure detection sensitivity and experimental reproducibility.
4. Customized Solutions
Adapts analysis workflows to sample characteristics and research objectives, meeting diverse requirements.
Applications
1. Protein Interaction Studies
By detecting the impact of methylation on binding interfaces, the stability and dynamic changes of protein complexes can be analyzed.
2. Transcriptional Regulation Mechanisms
The 4D-methylation proteomics service can be used to reveal the methylation status of transcription factors and related regulatory proteins, helping to understand gene expression regulatory networks.
3. Crosstalk of Protein Modifications
By studying the interplay between methylation and other post-translational modifications, multilayered regulatory mechanisms can be elucidated.
4. Functional Biomarker Discovery
The 4D-methylation proteomics service can be applied to identify methylation sites associated with specific physiological states, supporting molecular biomarker research.
FAQ
Q1: What Is the Difference between 4D Methylation Proteomics and Conventional Methylation Detection?
A1: The 4D platform combines ion mobility separation with high-resolution mass spectrometry, which can significantly improve the detection rate of low-abundance methylation sites in complex samples, reduce co-elution interference, and simultaneously enhance quantitative accuracy and reproducibility.
Q2: Can 4D Detection Distinguish between Mono-, Di-, and Tri-methylation?
A2: Yes. Through specific enrichment strategies and high-resolution fragment ion modes (such as ETD/EThcD), it can distinguish different methylation forms at the residue level and perform relative quantitative analysis.








