Histone Trimethylation Analysis Service
Based on a high-resolution liquid chromatography-tandem mass spectrometry (LC-MS/MS) platform, MtoZ Biolabs has launched the histone trimethylation analysis service to conduct systematic detection and quantitative research on histone trimethylation modifications. This service enables accurate identification of trimethylation sites on different lysine or arginine residues and analysis of their abundance changes under different conditions. Combined with bioinformatics methods, it can provide data on modification distribution patterns, differential comparisons, and functional annotations, helping researchers explore in depth the role of trimethylation in epigenetic regulation, gene expression, and cellular function studies, and providing reliable data support for related scientific and applied research.
Overview
Histone trimethylation refers to a type of post-translational modification in which three methyl groups are attached to lysine or arginine residues on histones, playing a key role in maintaining chromatin structure and regulating gene transcription. Trimethylation can occur on various lysine residues, including H3K4, H3K9, H3K27, H4K20, and H4K36, each with distinct regulatory functions. Histone trimethylation analysis can systematically detect and quantify trimethylation modifications at different sites, and is widely applied in epigenetics research, gene expression regulation, cell fate studies, and potential biomarker discovery, providing reliable support for understanding their biological functions.
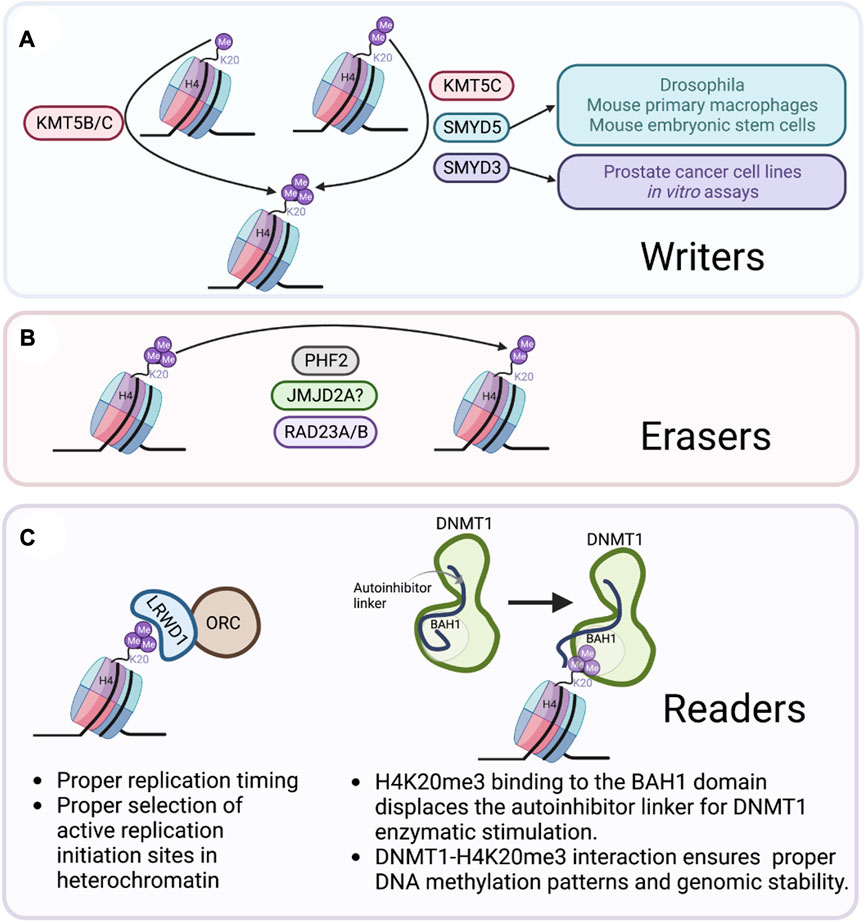
Agredo, A. et al. Frontiers in Genetics, 2023.
Figure 1. Writers, Erasers, and Readers of H4K20me3.
Analysis Workflow
1. Histone Extraction
Histones are isolated from cell or tissue samples.
2. Digestion and Modification Enrichment
Enzymatic digestion is performed under optimized conditions, and trimethylated peptides are captured using specific enrichment strategies.
3. LC-MS/MS Detection
Relying on a high-resolution liquid chromatography–tandem mass spectrometry platform, modification sites are identified and quantified.
4. Data Analysis
Combined with bioinformatics methods, results on modification distribution and differences are provided to support functional studies.
Sample Submission Suggestions
1. Sample Type and Quantity
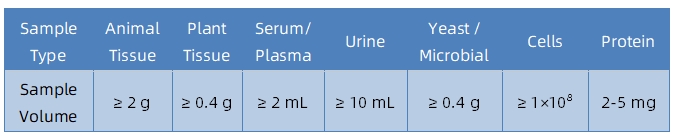
Note: Plasma should be collected using EDTA as an anticoagulant. Standard tissue or cell lysis buffers can be used during protein extraction.
2. Sample Transportation
Avoid repeated freeze-thaw cycles. Samples are recommended to be stored at -80°C and transported on dry ice to ensure low-temperature conditions throughout the process and prevent modification loss.
Note: For special samples or if a detailed submission plan is required, please contact MtoZ Biolabs technical staff in advance.
Service Advantages
1. High-Resolution Detection
Relying on advanced LC-MS/MS platforms, low-abundance trimethylation modification sites can be accurately identified.
2. Efficient Enrichment Strategy
Optimized peptide enrichment methods are applied to significantly improve modification detection rates and coverage.
3. High Data Quality
Standardized workflows and strict quality control ensure accurate and reliable analytical results.
4. Flexible Customized Solutions
Experimental plans can be tailored according to sample types and research objectives to meet diverse needs.
Applications
1. Epigenetics Research
Histone trimethylation analysis service can be used to analyze the role of trimethylation in chromatin conformation changes and nucleosome stability.
2. Cell Development Research
By analyzing trimethylation modifications during different developmental stages or cell differentiation, their dynamic changes can be explored.
3. Environmental Response Research
Histone trimethylation analysis service can be applied to study the variation patterns of trimethylation levels under stress, nutrient conditions, or external stimuli.
4. Potential Biomarker Discovery
By identifying trimethylation features associated with specific physiological or pathological states, it contributes to novel biomarker screening.
FAQ
Q1: Can Histone Trimethylation Be Analyzed in Combination with Other Histone Modifications?
A1: Yes. Trimethylation often works together with acetylation, monomethylation, or dimethylation to jointly regulate chromatin states. Combined analysis helps reveal the interactions between modifications and their complex regulatory mechanisms.
Q2: Do Different Trimethylation Sites Have Functional Differences?
A2: Yes. Trimethylation at different residues often has distinct regulatory roles. For example, H3K4me3 is usually associated with transcriptional activation, while H3K9me3 or H3K27me3 is more often related to gene silencing or repression. Therefore, site-specific analysis is essential.








