Ubiquitin Proteomics Service
MtoZ Biolabs offers systematic Ubiquitin Proteomics Service powered by Thermo Fisher Orbitrap Fusion, Q Exactive HF, and Orbitrap Lumos mass spectrometers combined with Nano-LC. With high sensitivity and resolution, our platform enables precise site mapping, enrichment of ubiquitinated peptides, quantitative comparison, and advanced bioinformatics analysis. Through integrated workflows and reproducible pipelines, we deliver comprehensive ubiquitinome profiling to support biomedical research, drug discovery, and agricultural biotechnology.
Overview
Ubiquitination is a key post-translational modification that regulates protein stability, degradation, trafficking, and signal transduction. By covalently attaching ubiquitin molecules to lysine residues of substrate proteins, cells control processes ranging from protein turnover via the proteasome to immune regulation and stress responses. Dysregulation of ubiquitination is closely linked to cancer, neurodegeneration, inflammation, and other major diseases, making ubiquitinome research vital for understanding disease mechanisms and developing therapeutic strategies.
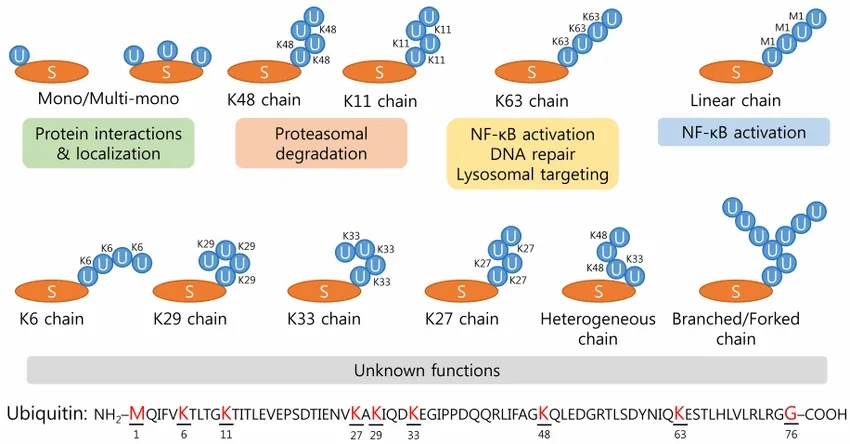
Park, C. et al. BMB Rep. 2014.
Figure 1. Figure 1. Various Types of Ubiquitination and the Specific Role of Ub Chain Linkages
Services at MtoZ Biolabs
MtoZ Biolabs provides a comprehensive suite of ubiquitinome services tailored to different research needs:
● Ubiquitination Site Identification Service
Accurate mapping of ubiquitination sites on substrate proteins to reveal modification patterns and regulatory hotspots.
● Ubiquitinated Peptide Enrichment Service
Selective enrichment of ubiquitinated peptides using affinity-based strategies to improve detection of low-abundance modifications.
● Quantitative Ubiquitylomics Service
Label-Free, TMT/iTRAQ, and DIA quantification methods provide dynamic comparisons of ubiquitination across different conditions.
● DIA-MS Ubiquitinome Analysis Service
Data-independent acquisition (DIA-MS) workflows deliver high-throughput and reproducible ubiquitinome profiling with broad coverage.
● Ubiquitylomics Data Analysis Service
Integrated bioinformatics including differential analysis, pathway enrichment, and protein network modeling for comprehensive data interpretation.
Analysis Workflow
1. Protein Extraction: Isolation of proteins from cells, tissues, serum, plasma, or exosome samples with quality control.
2. Enzymatic Digestion: Proteins are digested into peptides suitable for LC-MS/MS analysis.
3. Ubiquitinated Peptide Enrichment: DiGly (K-ε-GG) remnant antibody enrichment and complementary strategies are used to selectively capture ubiquitinated peptides.
4. Mass Spectrometry Detection: High-resolution Orbitrap instruments combined with Nano-LC for sensitive and accurate ubiquitinome analysis.
5. Data Analysis: Site identification, quantification, pathway annotation, and protein network modeling using mainstream bioinformatics software.
6. Report Delivery: Comprehensive report including site distribution, quantitative comparisons, pathway annotations, and visualized results.
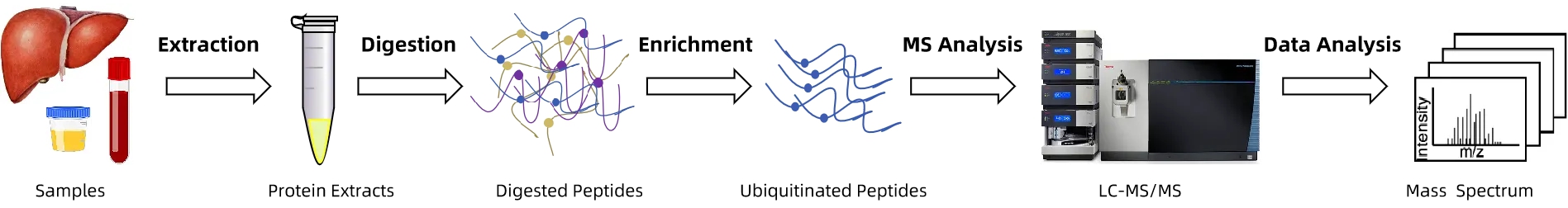
Figure.2 A Generic Workflow of Ubiquitin Proteomics Service
Why Choose MtoZ Biolabs?
MtoZ Biolabs delivers robust ubiquitin proteomics solutions with distinct advantages:
✅ Optimized enrichment strategies: DiGly remnant antibody capture combined with complementary methods ensures comprehensive ubiquitinome coverage.
✅ High throughput capability: Capable of profiling thousands of ubiquitination sites in a single experiment for large-scale studies.
✅ State-of-the-art instrumentation: Orbitrap and timsTOF platforms deliver superior resolution and sensitivity.
✅ Expert scientific team: Extensive experience in ubiquitin signaling and post-translational modification proteomics.
✅ Actionable data reporting: Clear, publication-ready datasets with advanced bioinformatics interpretation.
✅ Transparent pricing: One-time charge policy with no hidden fees.
Sample Submission Suggestions

Note: Provide details on sample collection and handling. If you need further details, our technical support team is happy to assist and provide comprehensive guidance on sample submission.
Applications
Ubiquitin proteomics provides powerful insights into cellular processes and disease mechanisms, with applications including:
Cancer research: Identifying ubiquitin-regulated oncogenic signaling pathways and drug resistance mechanisms.
Neurodegenerative disease studies: Exploring protein degradation and aggregation mediated by ubiquitination.
Drug discovery: Characterizing E3 ligase substrates and evaluating PROTAC-mediated degradation.
Cell signaling: Deciphering ubiquitin-driven regulation of signaling cascades.
Biomarker discovery: Detecting ubiquitination-based markers for diagnostics and precision medicine.
FAQ
Q1: At which sites does ubiquitination typically occur and how is it detected?
Ubiquitination primarily occurs on lysine residues, though non-lysine sites are occasionally observed. After trypsin digestion, ubiquitinated lysines carry a diglycine (diGly) remnant that serves as a unique signature, enabling site-specific enrichment with anti-K-ε-GG antibodies and LC-MS/MS detection.
Q2: What is the significance of different polyubiquitin chain linkages such as K48 and K63?
Chain linkage type dictates protein fate. K48-linked chains typically signal proteasomal degradation, while K63-linked chains mediate non-proteolytic functions including DNA repair, signal transduction, and endocytosis. Other linkages (K6, K11, K27, K29, K33, M1) confer distinct regulatory roles, actively studied in functional ubiquitinomics.
By integrating advanced enrichment strategies with high-resolution mass spectrometry, MtoZ Biolabs delivers robust and high-throughput results through our Ubiquitin Proteomics Service that enables large-scale and accurate exploration of ubiquitin signaling. Beyond ubiquitination, we provide a full portfolio of post-translational modification proteomics services, offering comprehensive datasets that accelerate research in biology, drug discovery, and precision medicine.








