M-LAC-Based Modified Peptide Enrichment Service
Post-translational modifications (PTMs) are crucial regulators of protein structure and function, affecting processes such as cell signaling, gene expression, and metabolic regulation. However, the low abundance and complex nature of modified peptides present significant analytical challenges. Multi-lectin affinity chromatography (M-LAC) has emerged as an effective enrichment method for selectively isolating glycopeptides and other modified biomolecules, leveraging the complementary binding properties of multiple lectins to achieve broad coverage and enhanced specificity.
MtoZ Biolabs provides a comprehensive M-LAC-Based Modified Peptide Enrichment Service designed to enrich, identify, and quantify glycosylated and other lectin-binding peptides with high precision. Our workflows integrate multi-lectin affinity chromatography, HILIC fractionation, and high-resolution LC-MS/MS analysis to deliver sensitive, reproducible, and biologically meaningful results. In addition, we combine lectin selection optimization with bioinformatics-based glycan profiling, enabling the identification of site-specific modifications and their biological implications across different sample types and research models. Our M-LAC-Based Modified Peptide Enrichment Service supports both targeted glycopeptide analysis and proteome-wide glycoproteomics, helping researchers explore the structural diversity of glycans, elucidate glycoprotein function, and uncover disease-associated glycosylation patterns.
Technical Principles
Multi-Lectin Affinity Chromatography (M-LAC) is based on the specific and reversible interactions between lectins and carbohydrate moieties on glycoproteins or glycopeptides. Lectins are a diverse class of carbohydrate-binding proteins, each recognizing distinct sugar motifs such as mannose, galactose, fucose, N-acetylglucosamine, or sialic acid. Commonly used lectins include Concanavalin A (ConA), Wheat Germ Agglutinin (WGA), Lens culinaris agglutinin (LCA) and Phaseolus vulgaris leucoagglutinin (PHA-L), Phaseolus vulgaris erythroagglutinin (PHA-E), and Sambucus nigra agglutinin (SNA). By immobilizing multiple lectins with complementary specificities on a solid support, M-LAC enables the simultaneous capture of a wide spectrum of glycosylated peptides and proteins within complex biological mixtures.
In this system, the stationary phase is composed of a matrix (commonly agarose or silica) covalently bound to the selected lectin mixture. When a peptide or protein sample is passed through the M-LAC column, glycopeptides interact with the lectins via non-covalent hydrogen bonding, van der Waals forces, and hydrophobic interactions between the lectin binding site and the glycan residues. Peptides that lack compatible carbohydrate structures are washed away using optimized buffer conditions.
Elution is achieved through competitive displacement with specific sugars (such as methyl α-D-mannopyranoside or N-acetylglucosamine) or by altering the ionic strength and pH of the buffer, thereby disrupting the lectin–carbohydrate interactions without damaging the glycan structure. The eluted fractions contain glycopeptides of diverse glycan compositions and linkage types, which can then be analyzed by high-resolution liquid chromatography–mass spectrometry (LC-MS/MS).
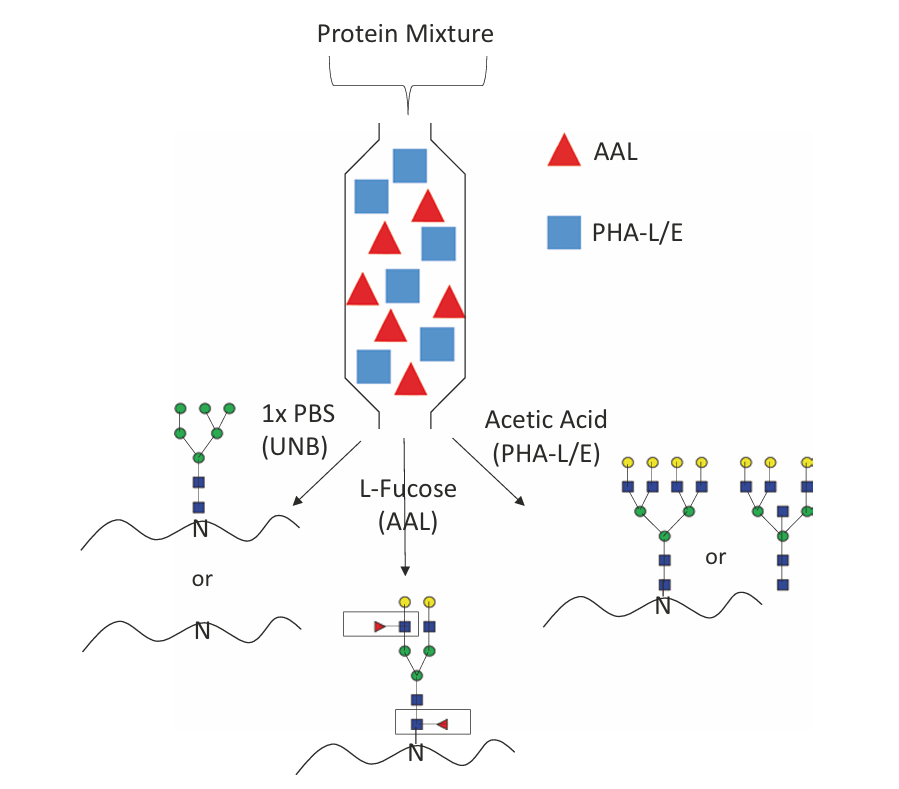
Totten, S. M. et al. Methods Mol Biol. 2017.
Figure 1. Multi-lectin Affinity Chromatography Schematic
Analysis Workflow
1. Protein Extraction and Digestion
Proteins are extracted from cells, tissues, or biofluids, reduced, alkylated, and digested into peptides while maintaining glycan integrity.
2. Lectin Selection and Column Preparation
Multiple lectins with complementary binding affinities are covalently immobilized on solid-phase supports to maximize glycopeptide capture efficiency.
3. Enrichment of Modified Peptides
The digested peptides are passed through the M-LAC column, allowing glycopeptides and other lectin-binding peptides to bind specifically, while unmodified peptides are washed away.
4. Elution and Fractionation
Bound glycopeptides are eluted using competitive sugars or specific buffer gradients, followed by fractionation when necessary for improved separation.
5. Mass Spectrometry Detection
High-resolution LC-MS/MS is used to identify peptide sequences, glycosylation sites, and glycan compositions with high confidence.
6. Bioinformatics Analysis and Data Interpretation
Advanced bioinformatics tools are employed for glycan structure annotation, site-specific quantification, and biological pathway mapping.
Sample Submission Suggestions
1. Sample Type: Cell lysates, tissue extracts, serum/plasma, or purified glycoproteins.
2. Minimum Requirement: 200–500 µg of total protein per sample.
3. Storage Conditions: Freeze samples at -80°C and ship on dry ice to prevent glycan degradation.
*Note: For other sample types or low-input experiments, please contact MtoZ Biolabs for customized preparation guidance.
Service Advantages
✅ Broad Glycan Coverage
Simultaneous use of multiple lectins ensures enrichment of diverse glycopeptides, capturing a wide range of glycan structures.
✅ High Specificity and Sensitivity
Optimized multi-lectin combinations and refined washing protocols minimize background interference and maximize binding efficiency.
✅ Preservation of Glycan Integrity
Gentle elution conditions maintain native glycan structures, allowing accurate downstream characterization.
✅ Comprehensive Workflow
From sample preparation to data interpretation, we provide a complete end-to-end solution for glycopeptide enrichment and analysis.
✅ Customizable Solutions
Lectin combinations and enrichment conditions are tailored to specific biological systems and research objectives.
✅ Expert Team Support
Our experienced scientists in glycoproteomics and mass spectrometry provide professional guidance and data interpretation throughout the project.
Applications
1. Glycoproteomics Research
Characterize global glycosylation patterns across biological systems to understand cellular communication and protein trafficking.
2. Disease Biomarker Discovery
Identify glycosylation alterations associated with cancer, neurodegeneration, inflammation, and immune disorders.
3. Therapeutic Glycoprotein Characterization
Analyze glycan occupancy, branching, and composition to ensure the quality and efficacy of biopharmaceuticals.
4. Functional Proteomics
Study the regulatory effects of glycosylation on protein stability, folding, and receptor binding.
5. Comparative and Quantitative Studies
Quantify glycan modifications under different physiological or pathological conditions to reveal functional correlations.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
MtoZ Biolabs' M-LAC-Based Modified Peptide Enrichment Service bridges analytical precision with biological insight, providing a comprehensive path to decode the molecular language of glycosylation. Contact us to explore tailored solutions for your research.








