Protein Ubiquitination Analysis Service
Based on a high-resolution mass spectrometry platform and specific ubiquitinated peptide enrichment technology, MtoZ Biolabs has launched the protein ubiquitination analysis service which enables systematic identification and quantitative analysis of ubiquitination sites in protein samples. This service comprehensively analyzes the site distribution, modification levels, and dynamic changes of ubiquitination, and integrates database searches and bioinformatics tools to deliver differential analysis and signaling pathway annotation. The final results include not only accurate ubiquitination site identification but also quantitative data and functional pathway information, providing researchers with reliable evidence for elucidating protein degradation, signal regulation, and disease mechanisms.
Overview
Protein ubiquitination refers to the process in which the small protein ubiquitin is covalently attached to target proteins through the cascade action of ubiquitin-activating enzyme, ubiquitin-conjugating enzyme, and ubiquitin ligase. It is one of the most important post-translational modifications regulating protein degradation, signal transduction, and cellular homeostasis. The analytical principle mainly relies on high-resolution mass spectrometry technology, combined with specific enrichment strategies, to achieve accurate identification and quantification of ubiquitination sites in protein samples, thereby revealing the role of ubiquitination in regulating protein fate and signal networks. This method is widely applied in disease mechanism research, drug target screening, and cellular function analysis, providing essential evidence for a deeper understanding of cellular homeostasis and disease development.
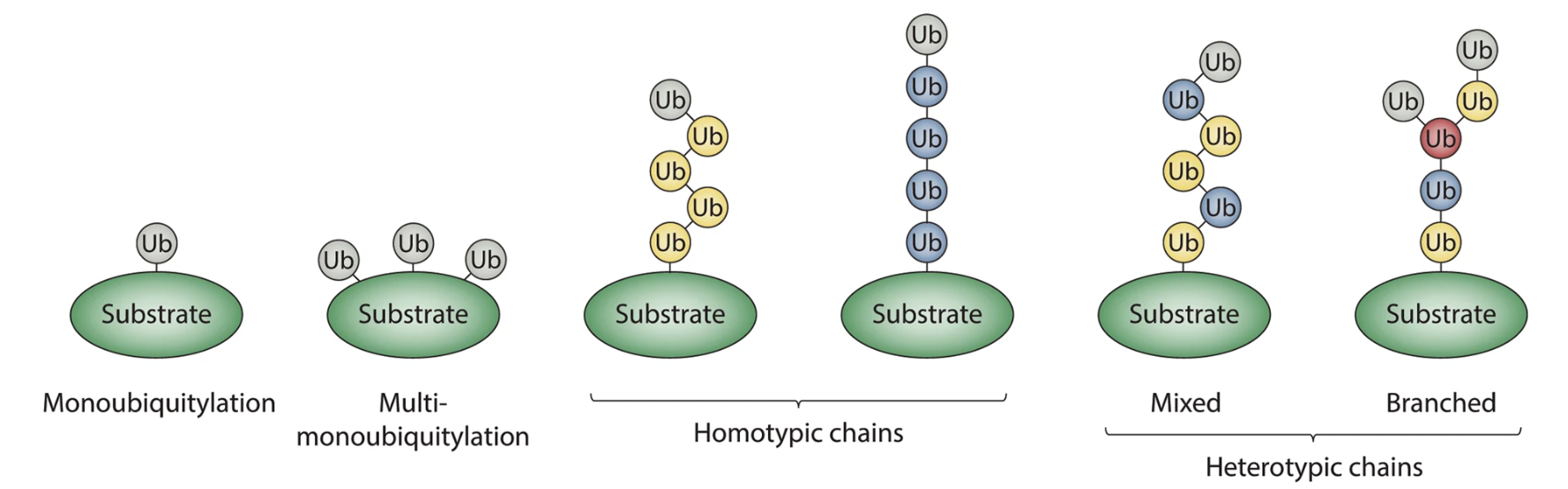
French, M. E. et al. Cell Discov. 2021.
Figure 1. Classification of Ubiquitin Modifications.
Services at MtoZ Biolabs
1. Target Protein Ubiquitination Analysis
MtoZ Biolabs performs qualitative and site-specific analysis of ubiquitination on designated target proteins, including distinguishing mono-ubiquitination, multi-ubiquitination, and linkage types such as K48 and K63. With a high-resolution LC-MS/MS platform, we accurately identify modification site changes and support researchers in evaluating how ubiquitination influences protein stability, degradation, or regulatory behavior.
2. Ubiquitinomics Analysis
MtoZ Biolabs applies the K-ε-GG antibody enrichment strategy combined with advanced mass spectrometry systems to achieve proteome-wide identification and quantification of ubiquitinated peptides. This analysis delineates large-scale ubiquitination networks and reveals global modification patterns associated with regulatory pathways, stress responses, and protein fate.
Analysis Workflow
1. Sample Preparation
Protein extraction and quantification of samples such as cells, tissues or body fluids to ensure uniform quality.
2. Protein Digestion
Proteins are digested into peptides to facilitate subsequent selective enrichment and detection of ubiquitination modifications.
3. Ubiquitinated Peptide Enrichment
K-ɛ-GG antibody or affinity chromatography methods are used to enrich ubiquitinated peptides, improving detection sensitivity and accuracy.
4. Mass Spectrometry Detection
Based on a high-resolution LC-MS/MS platform, the enriched peptides are subjected to precise qualitative and quantitative analysis.
5. Data Analysis
Databases and bioinformatics tools are employed to annotate ubiquitination sites, providing distribution features, differential levels, and related signaling pathway information.
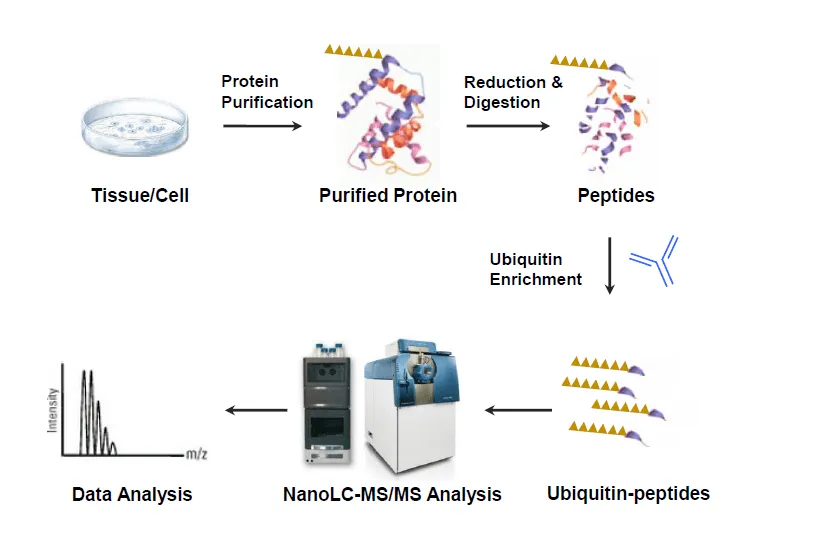
Figure 2. The Workflow of Protein Ubiquitination Analysis.
Sample Submission Suggestions
1. Sample Type and Quantity
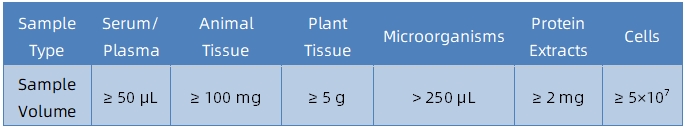
2. Sample Storage
Samples should be stored at low temperatures (such as -80°C frozen) to avoid repeated freeze–thaw cycles, which may cause protein degradation and loss of ubiquitination modifications.
3. Sample Transportation
During transportation, it is recommended to use dry ice or cold chain conditions to ensure that the samples remain intact and the modifications stable before arriving at the analytical platform.
Service Advantages
1. High-Sensitivity Detection
Relying on a high-resolution LC-MS/MS platform and optimized experimental workflows, low-abundance ubiquitination sites can be accurately detected in complex samples, ensuring data reliability.
2. Specific Enrichment
By combining specific antibodies with affinity chromatography and other enrichment strategies, ubiquitinated peptides are effectively separated, improving the signal-to-noise ratio and detection accuracy.
3. Professional Team Support
Experiments and data analysis are conducted by experts with extensive experience in protein modification research and mass spectrometry analysis, ensuring standardized operations and trustworthy results.
4. Customized Solutions
According to different research goals and drug development needs, the depth and scope of analysis can be flexibly adjusted to provide personalized detection and interpretation strategies.
Applications
1. Drug Effect Evaluation
By detecting changes in ubiquitination levels before and after drug treatment, this approach is used to verify the regulatory effects of drugs on their targets and downstream signaling pathways.
2. Protein Degradation Pathway Research
Protein ubiquitination analysis service can be used to systematically analyze the modification characteristics of target proteins in the ubiquitin–proteasome pathway, helping to explore protein stability and degradation mechanisms.
3. Cellular Signal Regulation Analysis
Through the identification and quantification of ubiquitination sites, the role of ubiquitination in signal transduction and the regulation of cellular functions can be revealed.
4. Biomarker Discovery
Protein ubiquitination analysis service can be used to identify ubiquitination sites associated with specific physiological or pathological states, providing a basis for the screening of potential diagnostic or prognostic biomarkers.
FAQ
Q1: Why Is Mass Spectrometry Required for Protein Ubiquitination Detection Instead of Traditional Methods?
A1: Ubiquitination is a low-abundance and highly heterogeneous modification, and traditional methods cannot provide a comprehensive analysis. Mass spectrometry combined with specific enrichment can simultaneously identify and quantify thousands of ubiquitination sites at a global level, offering systematic and high-resolution data.
Q2: Does This Service Support Comparisons Across Different Experimental Conditions or Treatment Groups?
A2: Yes. Through quantitative analysis strategies, ubiquitination levels can be compared across multiple sample groups, revealing modification changes under different treatments or biological states.
Q3: Can Ubiquitination Analysis Be Combined with Studies of Other Post-Translational Modifications?
A3: Yes. This service can be integrated with analyses of phosphorylation, acetylation, and other modifications, helping to construct a multidimensional protein modification network and reveal more comprehensive regulatory mechanisms.








