Histone PTM Data Analysis Service
MtoZ Biolabs' histone PTM data analysis service focuses on the systematic interpretation of mass spectrometry data related to histone post-translational modifications (PTMs). The service covers data preprocessing, modification site identification, quantitative comparison, and functional annotation, generating visual outputs such as heatmaps, volcano plots, principal component analysis (PCA) plots, and pathway enrichment charts. Through multidimensional statistical and bioinformatics analyses, it helps researchers accurately identify key modification changes and uncover epigenetic regulatory patterns and potential functional associations.
Overview
Histone post-translational modifications (PTMs) refer to various chemical modifications that occur on histones after translation, which regulate chromatin conformation and gene transcription activity, playing critical roles in epigenetic regulation. With advancements in mass spectrometry technologies, histone modification research has entered a high-throughput and multidimensional era, making data interpretation a key step in elucidating regulatory mechanisms. Histone PTM data analysis systematically processes mass spectrometry data to achieve modification site identification, quantitative comparison, and functional annotation, providing in-depth insights for studies of epigenetic regulation, cell fate determination, and molecular network characterization.
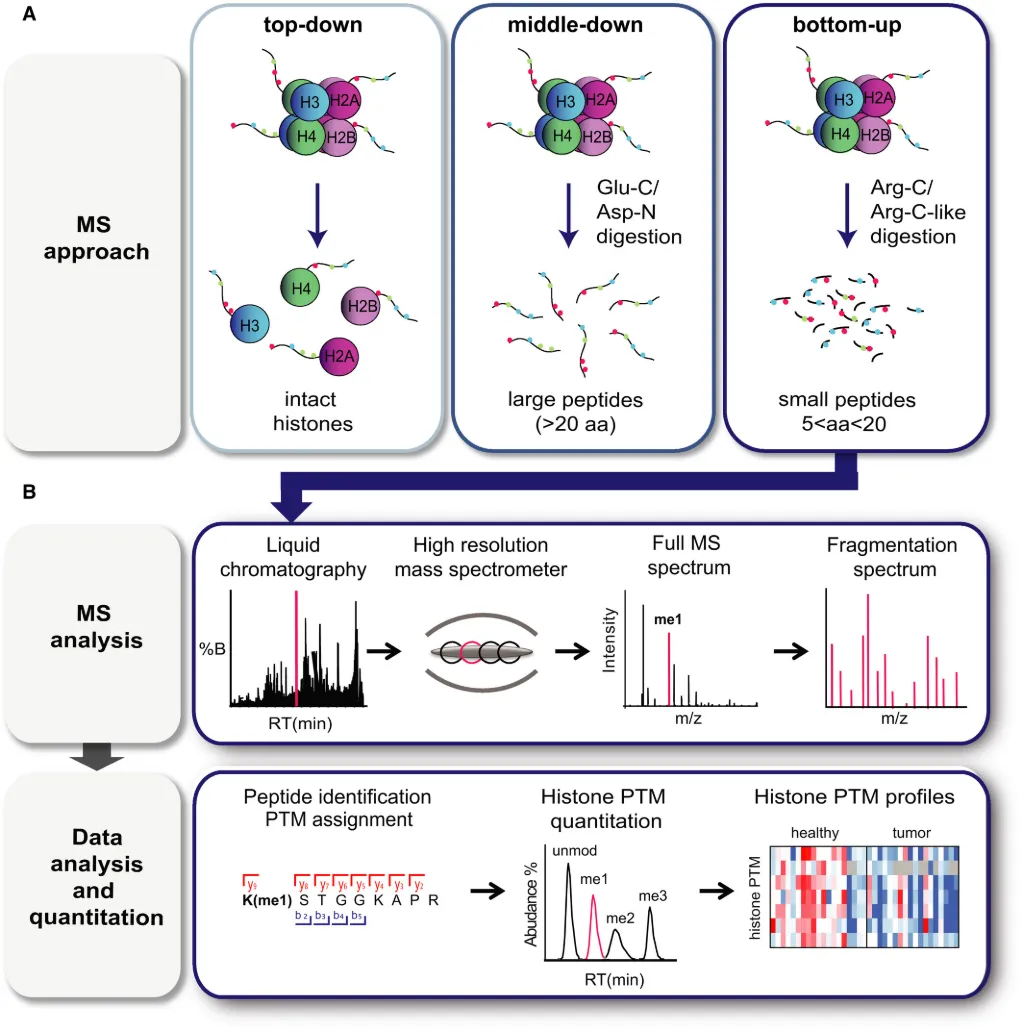
Noberini, R. et al. FEBS J. 2022.
Figrue 1. MS-Based Analysis of Histone PTMs.
Service Advantages
1. Multidimensional Data Interpretation
Integrates qualitative, quantitative, and statistical analyses to systematically elucidate histone modification patterns and dynamic trends.
2. Efficient Data Visualization
Generates multiple graphical representations such as volcano plots, heatmaps, and PCA plots, providing an intuitive display of modification differences and sample distribution.
3. Comprehensive Functional Annotation
Utilizes GO and KEGG databases to perform pathway enrichment and biological function analysis of histone modifications.
4. Standardized Analytical Workflow
Employs rigorous data processing and quality control procedures to ensure analytical accuracy, consistency, and reproducibility.
Applications
1. Epigenetic Regulation Research
The histone PTM data analysis service can be applied to elucidate the role of histone modification patterns in chromatin structure modulation and transcriptional activation.
2. Differential Modification Screening
Through quantitative and statistical analyses, it identifies PTMs that exhibit significant changes under different experimental or physiological conditions.
3. Multi-Omics Integration
The histone PTM data analysis service can be combined with transcriptomic, metabolomic, or protein interaction data to construct a comprehensive regulatory network.
4. Potential Biomarker Discovery
By comparing modification differences across samples, it assists in identifying potential functional or epigenetic regulatory biomarkers.
Deliverables
1. Raw Data and Quality Assessment
Provides original mass spectrometry or sequencing data files accompanied by a quality control report to ensure data integrity and usability.
2. PTM Site Identification Results
Delivers a high-confidence list of histone modification sites, including residue-level localization and modification types (e.g., acetylation, methylation states).
3. Quantitative and Differential Analysis
Includes standardized quantitative data, fold-change analysis, and statistical significance evaluation (p-value/FDR).
4. Functional and Pathway Annotation
Performs enrichment and pathway visualization analyses based on GO, KEGG, and Reactome databases, presenting biologically relevant insights.
5. Visualization and Comprehensive Report
Provides multidimensional data visualizations such as volcano plots, heatmaps, and PCA plots, along with a detailed analytical report and biological interpretation.








