Protein SUMOylation Analysis Service
Based on a high-resolution LC-MS/MS mass spectrometry platform and specific enrichment technology, MtoZ Biolabs has launched the protein SUMOylation analysis service which can perform systematic analysis of SUMOylation modification sites in protein samples. This service enables accurate identification and quantification of SUMOylation sites, providing modification distribution, abundance changes, and potential functional information. The final results not only deliver high-confidence modification site data but can also be combined with bioinformatics analysis to reveal their role in cellular function regulation and molecular networks, offering reliable support for scientific research projects.
Overview
Protein SUMOylation refers to the process in which small ubiquitin-like modifier (SUMO) proteins are covalently attached to lysine residues of target proteins under the catalysis of E1 activating enzyme, E2 conjugating enzyme, and E3 ligase. It is a reversible post-translational modification. SUMOylation can affect protein stability, localization, and activity, playing important roles in transcriptional regulation, DNA repair, cell cycle, and stress response. Its analysis mainly relies on high-resolution mass spectrometry combined with specific enrichment, enabling precise identification and quantification of modification sites, and revealing modification patterns and functional characteristics. This method has been widely applied in signal pathway research, molecular mechanism analysis, and cellular functional network construction, providing data support for understanding complex biological processes.
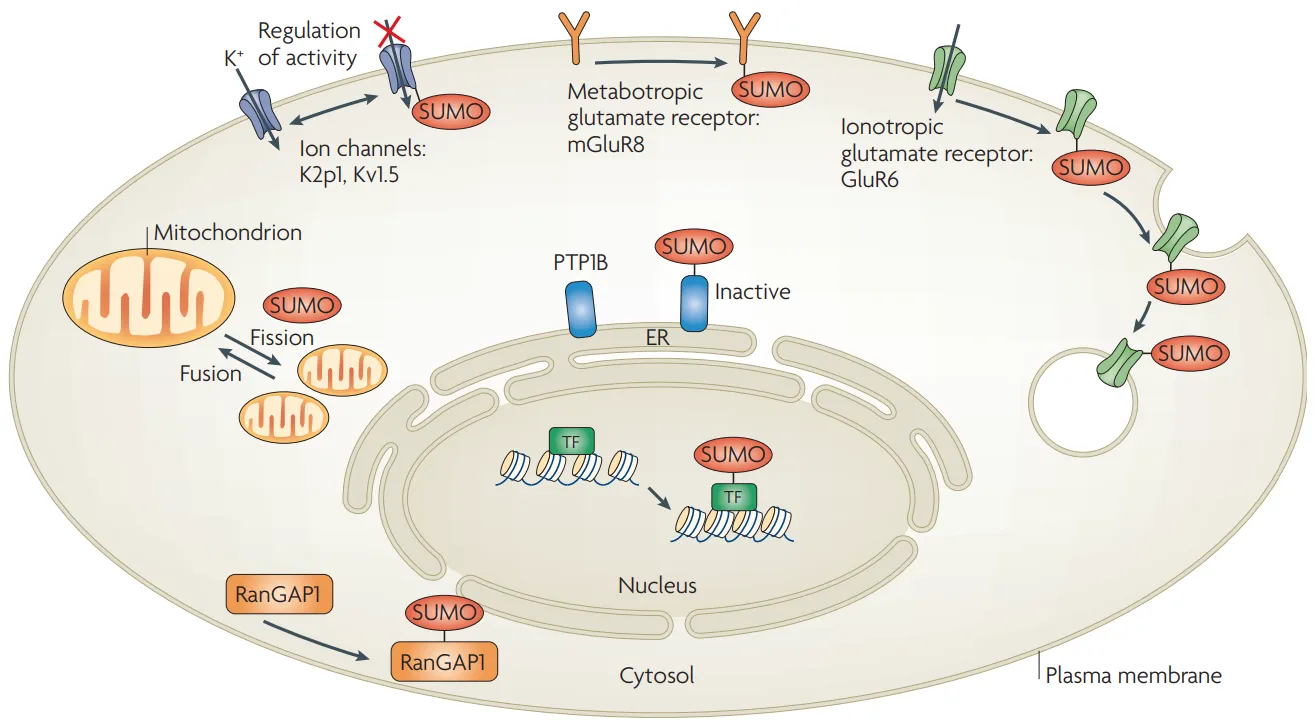
Geiss-Friedlander, R. et al. Nat Rev Mol Cell Biol, 2007.
Figure 1. Protein Sumoylation Identification.
Services at MtoZ Biolabs
1. Target Protein SUMOylation Analysis
MtoZ Biolabs performs SUMOylation modification identification for client-designated target proteins, including determination of modification sites, SUMO chain types, and alteration patterns under different experimental conditions. Powered by a high-resolution LC-MS/MS platform, we analyze how SUMOylation affects target protein stability, conformation, or regulatory properties, providing key evidence for mechanistic studies.
2. SUMOylation Proteomics Analysis
MtoZ Biolabs applies K-ε-SUMO-specific antibody enrichment combined with large-scale mass spectrometry to systematically identify and quantify extensive SUMOylated peptides in complex samples. This analysis enables the construction of a global SUMOylation map, revealing regulatory network changes across conditions and providing comprehensive data for understanding the role of SUMOylation in cellular responses and protein function regulation.
Analysis Workflow
1. Sample Preparation
Perform protein extraction and quantification from cell, tissue, or body fluid samples to ensure sample quality and uniformity.
2. Protein Digestion
Digest proteins into peptides to facilitate the subsequent detection of SUMOylation modifications.
3. SUMOylated Peptide Enrichment
Use anti-SUMOylation antibodies or affinity chromatography methods to selectively enrich SUMOylated peptides, improving detection sensitivity and specificity.
4. Mass Spectrometry Detection
Based on a high-resolution LC-MS/MS platform, conduct precise identification and quantification of the enriched products.
5. Data Analysis
Combine databases and bioinformatics tools to annotate SUMOylation sites, providing modification distribution, abundance changes, and potential functional information.
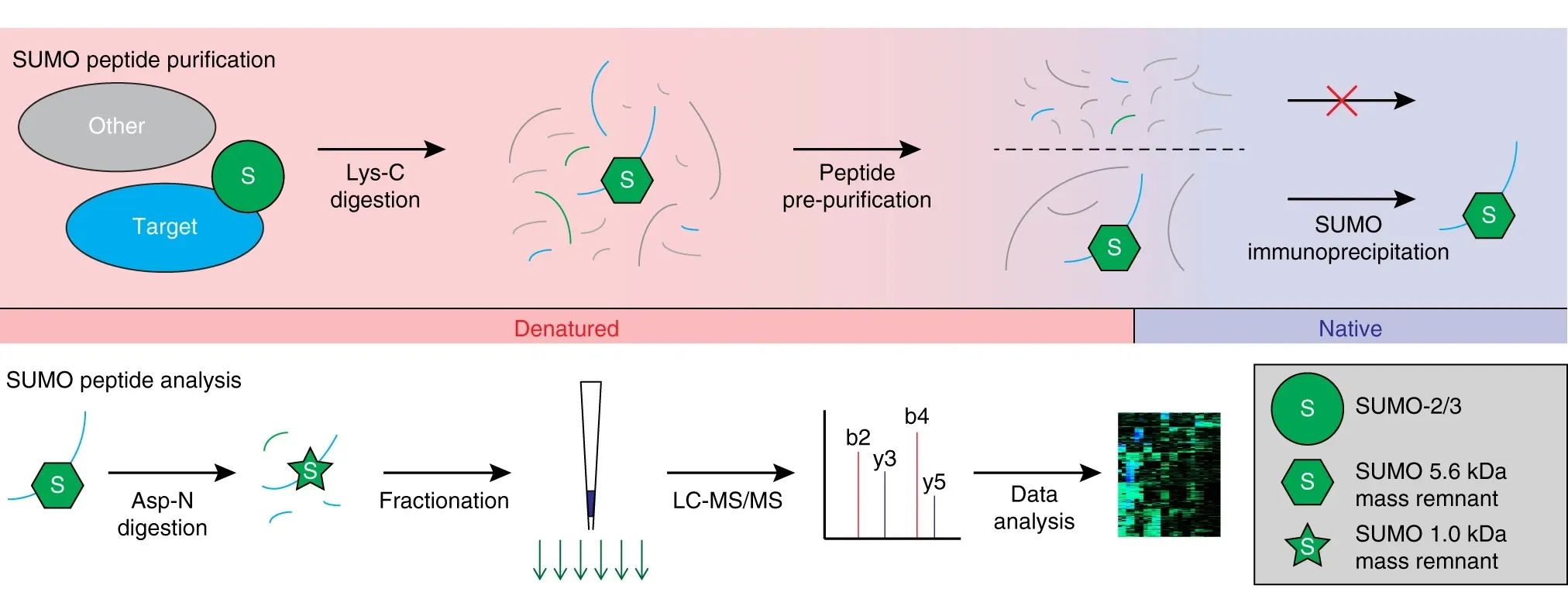
Hendriks, I A. et al. Nature Communications, 2018.
Figure 2. The Workflow of Protein Sumoylation Analysis.
Sample Submission Suggestions
1. Sample Type and Quantity
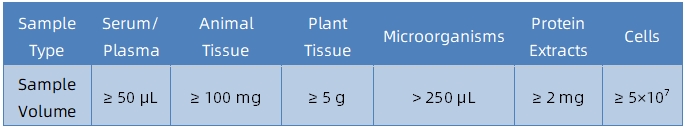
2. Sample Storage
Samples should be stored at low temperatures (such as at -80°C) to avoid repeated freeze-thaw cycles, preventing protein degradation and the loss of SUMOylation modifications.
3. Sample Transportation
During transportation, it is recommended to use dry ice or cold chain conditions along with sealed packaging to ensure the integrity and stability of samples before they reach the testing platform.
Service Advantages
1. High Sensitivity
Relying on a high-resolution LC-MS/MS platform, low-abundance SUMOylation sites can be accurately detected in complex backgrounds, ensuring reliable results.
2. Specific Enrichment
By combining SUMOylated peptide-specific antibodies with affinity chromatography technology, the signal-to-noise ratio and detection accuracy are effectively improved.
3. Professional Team
Experiments and data analysis are conducted by experts with extensive experience in mass spectrometry and protein modification research, ensuring standardized procedures and credible results.
4. Customized Solutions
According to different research needs, the detection range and depth can be flexibly adjusted to provide targeted SUMOylation analysis support.
Applications
1. Cellular Function Research
The protein SUMOylation analysis service can be used to evaluate its impact on protein stability, localization, and activity, supporting the exploration of cellular functions and regulatory mechanisms.
2. Epigenetics Research
It is used to study the role of SUMOylation in transcriptional regulation and chromatin remodeling, helping to understand gene expression regulatory patterns.
3. Signal Pathway Research
The protein SUMOylation analysis service can be applied to analyze its role in the regulation of key cellular signaling pathways.
4. Protein Interaction Network
By combining SUMOylation site and quantitative information, it explores its regulatory role in protein-protein interaction networks, constructing a more complete cellular molecular map.
FAQ
Q1: Why Does Protein SUMOylation Detection Rely on Mass Spectrometry Technology?
A1: SUMOylation is a low-abundance and complex post-translational modification, and traditional methods are difficult to comprehensively analyze it. High-resolution mass spectrometry combined with enrichment strategies can simultaneously identify and quantify a large number of SUMOylation sites, providing higher sensitivity and accuracy.
Q2: What Quantitative Strategies Are Available?
A2: Common methods include label-free quantification, isotope labeling (such as TMT and iTRAQ), and DIA analysis. The appropriate quantification approach can be selected according to research needs.
Q3: What Are the Main Limitations of SUMOylation Analysis?
A3: For extremely low-abundance or unstable modification sites, detection may have sensitivity limitations. In addition, data analysis relies on databases and algorithms, and annotations of certain novel or unknown modifications still require cautious interpretation.








