Protein Acetylation Analysis Service
Based on an advanced high-resolution LC-MS/MS mass spectrometry platform, MtoZ Biolabs has launched the protein acetylation analysis service which enables systematic identification and quantification of acetylation sites in protein samples. This service combines specific enrichment technology and strict experimental procedures to not only analyze the site distribution and modification level of acetylation, but also reveal its dynamic changes and functional characteristics. The final output includes precise identification results of acetylation sites, quantitative difference information, and related pathway annotations, providing researchers with reliable data support for gaining deeper insights into cellular functional regulation and protein modification networks.
Overview
Protein acetylation refers to the process in which an acetyl group is attached to the lysine residues of proteins under the catalysis of acetyltransferases, and it is one of the most common post-translational modifications. It can influence protein stability, conformation, and interactions, thereby playing an important role in gene expression, metabolic regulation, and signal transduction. Protein acetylation analysis relies on high-resolution mass spectrometry technology, combined with specific enrichment or antibody-based recognition methods, to achieve precise identification and quantification of acetylation sites, revealing the distribution characteristics and dynamic changes of the modification. This approach has been widely applied in signal pathway research, epigenetics, and cellular function studies, providing reliable data support for a deeper understanding of cellular regulatory networks.
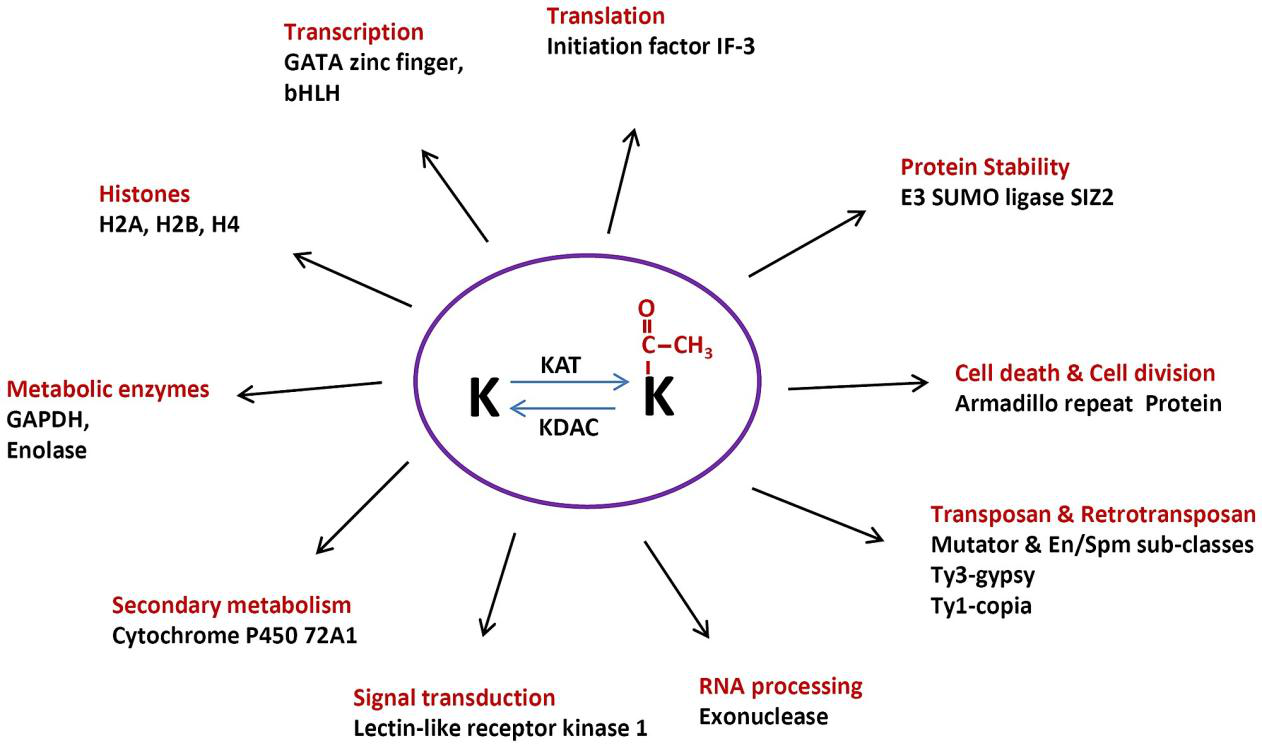
Nallamilli, B. R. R. et al. PloS one, 2014.
Figure 1. Multiple Regulatory Functions of Protein Acetylation.
Services at MtoZ Biolabs
1. Target Protein Acetylation Analysis
MtoZ Biolabs performs site-specific acetylation detection on designated target proteins, enabling precise identification of key amino acid residues that undergo acetylation and comparison of modification changes under different experimental conditions. Leveraging a high-resolution LC-MS/MS platform, we help researchers clarify how acetylation influences target protein structure and regulatory characteristics.
2. Acetylation Proteomics Analysis
MtoZ Biolabs combines anti-acetylation antibody enrichment with mass spectrometry-based proteomics to achieve systematic identification and quantification of acetylation modifications across the proteome. This analysis captures large numbers of acetylated peptides, reveals distribution patterns across diverse biological contexts, and provides a comprehensive data foundation for mechanism-focused research.
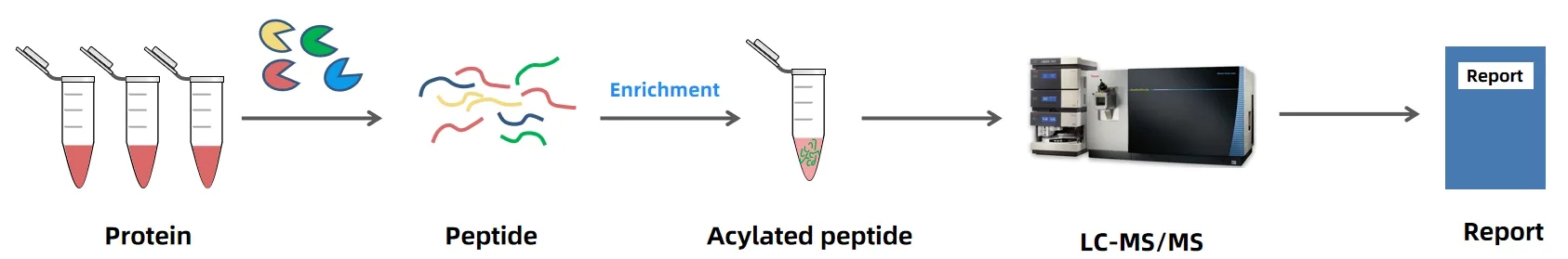
Analysis Workflow
1. Sample Preparation
Protein extraction and quantification of samples such as cells, tissues, or body fluids to ensure uniform quality.
2. Protein Digestion
Proteins are digested into peptides to facilitate the detection and analysis of acetylation modifications.
3. Acetylated Peptide Enrichment
Specific antibodies or affinity chromatography methods are used to selectively enrich acetylated peptides, improving detection sensitivity.
4. Mass Spectrometry Detection
Enriched peptides are analyzed with high precision identification and quantification using a high-resolution LC-MS/MS platform.
5. Data Analysis
Databases and bioinformatics tools are applied to annotate acetylation sites, providing distribution patterns, modification levels, and potential functional information.
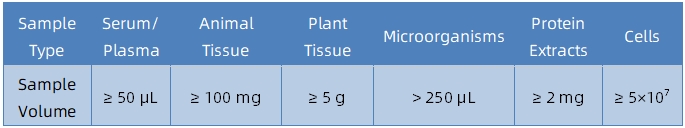
Sample Submission Suggestions
1. Sample Type and Quantity
2. Sample Storage
Samples should be stored under low-temperature conditions (such as -80°C freezing) to avoid repeated freeze-thaw cycles, preventing protein degradation and loss of acetylation modifications.
3. Sample Transportation
During transportation, dry ice or cold-chain conditions should be used to ensure that the samples maintain stability and integrity before reaching the analytical platform.
Service Advantages
1. High Sensitivity
With a high-resolution LC-MS/MS platform, low-abundance acetylation sites can be accurately detected, ensuring reliable results.
2. Specific Enrichment
By combining antibodies or affinity chromatography methods, acetylated peptides are effectively separated, improving the signal-to-noise ratio and detection accuracy.
3. Professional Team
Experiments and data analysis are carried out by experts with extensive experience in mass spectrometry and protein modification research, ensuring standardized procedures and credible results.
4. Customized Solutions
According to research needs, the analysis depth and scope can be flexibly adjusted to provide targeted data support.
Applications
1. Signal Pathway Research
Through acetylation site analysis, the activation state and regulatory mechanisms of key signaling pathways can be revealed.
2. Epigenetics Research
Used to analyze acetylation modifications of histones and other nuclear proteins, exploring their role in gene expression regulation.
3. Cell Function Analysis
Detection of acetylation in cell cycle, metabolic regulation, and molecular interactions supports functional studies.
4. Biomarker Discovery
By systematically analyzing acetylation sites, potential phenotype-related or functional biomarkers can be identified.
FAQ
Q1: Does the Service Support Large-Scale Batch Sample Analysis?
A1: Yes. This service supports parallel detection of multiple experimental conditions or treatment groups, allowing comparison of acetylation levels and helping to analyze dynamic changes.
Q2: Will Protein Acetylation and Other Modifications Interfere with Detection?
A2: In complex samples, acetylation often coexists with phosphorylation, methylation, and other modifications. High-resolution mass spectrometry can distinguish different modifications, but it requires advanced data analysis and accurate model matching.
Q3: Can Low-Abundance Acetylation Modifications Be Detected?
A3: Yes. By applying enrichment techniques such as IMAC and anti-acetylation antibodies, combined with a high-resolution LC-MS/MS platform, low-abundance acetylation sites can be accurately captured even in complex samples.








