SUMOylation Site Identification Service
Accurate identification of SUMOylation sites is key to understanding the biological functions of SUMO modification. However, due to the low abundance, reversibility, and dynamic nature of SUMOylation, traditional proteomic analyses often fail to directly detect SUMO-modified sites. By combining highly sensitive enrichment strategies with high-resolution mass spectrometry (MS), researchers can precisely capture and localize SUMOylation events within complex protein mixtures, providing deeper insights into the regulatory mechanisms of cellular signaling networks.
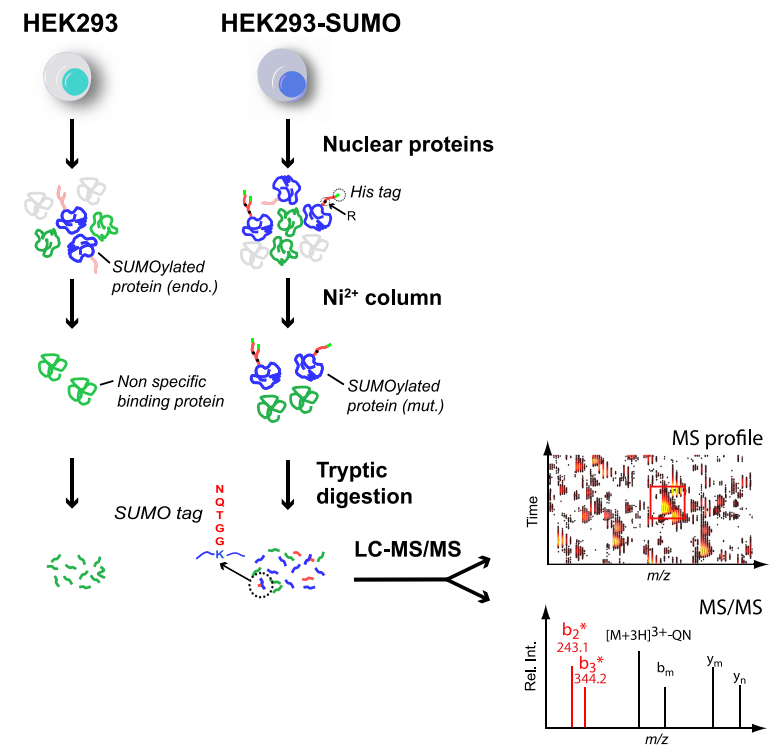
Galisson, F. et al. Mol Cell Proteomics. 2011.
Figure 1. SUMOylation Site Identification Workflow
Relying on advanced mass spectrometry platforms and extensive PTM research experience, MtoZ Biolabs offers a comprehensive SUMOylation site identification service that covers sample preparation, SUMOylated peptide enrichment, MS detection, and bioinformatic analysis to help researchers systematically explore the regulatory roles of SUMOylation in cellular physiology and pathology.
What is SUMOylation Modification
Post-translational modifications (PTMs) are essential biological mechanisms that regulate protein function and signaling pathway activity. Among them, SUMOylation is a modification in which a small ubiquitin-like modifier (SUMO) protein is covalently attached to the lysine residues of target proteins. This modification can influence protein stability, conformation, subcellular localization, and protein–protein interactions, playing crucial roles in processes such as DNA damage repair, chromatin remodeling, cell cycle control, and stress response.
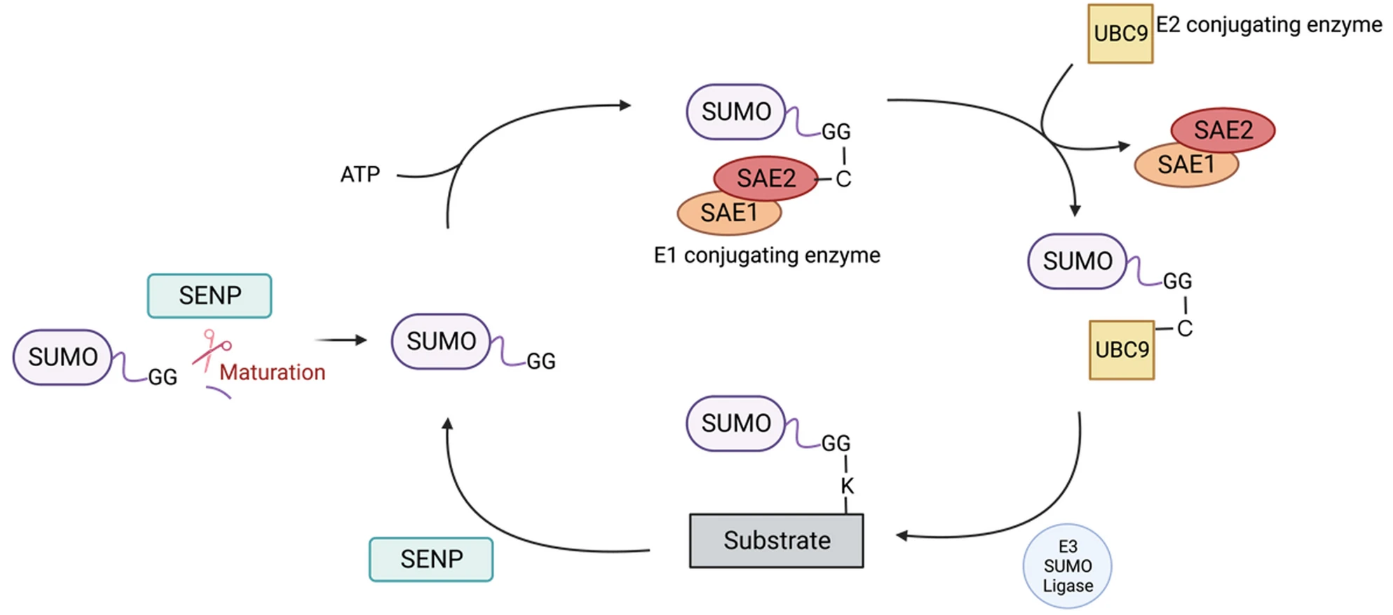
Huang, CH. et al. J Biomed Sci. 2024.
Services at MtoZ Biolabs
1. Target Protein SUMOylation Site Identification
MtoZ Biolabs provides Target Protein SUMOylation Site Identification focused on overcoming key challenges in studying SUMOylation on specific proteins, including targeted protein enrichment, capture of low-abundance modifications, and site-specific identification. Through immunoprecipitation, tag-based isolation, or anti-SUMO affinity enrichment, we selectively isolate the protein of interest from complex biological samples and perform high-resolution LC MS MS to precisely determine the amino acid residues modified by SUMO.
This service defines whether a specific protein is SUMOylated, identifies the exact modification sites, and evaluates how SUMOylation changes under different stimuli or mutational conditions, supporting structural studies, pathway analysis, and mechanistic validation.
2. SUMOylation Proteomics Site Identification
MtoZ Biolabs provides SUMOylation Proteomics Site Identification for large-scale, proteome-wide characterization and quantification of SUMOylation sites. By integrating anti-SUMO enrichment, SUMO-remnant peptide capture, and multidimensional peptide fractionation strategies such as HILIC and SCX with high-resolution LC MS MS and multiple fragmentation modes, we achieve deep profiling of hundreds to thousands of SUMOylation sites.
Supported by comprehensive bioinformatics analysis, this service reveals global SUMOylation patterns, dynamic regulation, site occupancy, and pathway associations, delivering systematic insights into signaling regulation, stress responses, disease mechanisms, and systems biology.
Analysis Workflow
1. Protein Extraction and Digestion
Optimized lysis and proteolysis strategies ensure efficient extraction and integrity of SUMOylated proteins.
2. SUMOylated Peptide Enrichment
Multiple enrichment methods, including antibody-based, affinity tag-based, and peptide-level approaches, enhance the detection sensitivity of SUMOylated peptides.
3. LC-MS/MS Analysis
High-resolution mass spectrometers (Orbitrap or Q Exactive series) provide precise detection and site-specific characterization of SUMOylated peptides.
4. Data Processing and Site Identification
Using professional software, SUMOylation sites are reliably identified and statistically validated.
5. Quantification and Functional Annotation (Optional)
Label-free, TMT, or SILAC quantification strategies are available, combined with GO and KEGG pathway analysis to reveal SUMO-related regulatory networks.
Service Advantages
1. Multi-tier Enrichment Strategies
Our SUMOylation site identification service integrates antibody-based capture, SUMO-tag affinity purification, and peptide-level enrichment approaches. These can be flexibly combined according to sample properties to enhance detection of low-abundance SUMOylated peptides.
2. High-resolution Mass Spectrometry Platform
Utilizing state-of-the-art Orbitrap and Q-TOF systems with advanced fragmentation methods (HCD, ETD, and EThcD), we achieve high-accuracy sequencing and precise localization of SUMOylation sites.
3. Expert Bioinformatics Analysis
Our experienced bioinformatics team provides comprehensive PTM data interpretation, including site annotation, pathway enrichment, and regulatory network analysis, offering biological context to SUMOylation findings.
4. Flexible and Customizable Solutions
We offer customizable analysis options including qualitative site identification, quantitative profiling, and multi-PTM integration, tailored to fit specific research needs.
Applications
1. Protein functional regulation and signaling pathway studies
2. Cell cycle and DNA damage repair mechanisms
3. Stress response and metabolic regulation research
4. Nuclear protein complex assembly and disassembly analysis
5. Novel target discovery and drug mechanism exploration
Sample Submission Suggestions
1. Sample Type: Cells, tissues, or purified protein samples
2. Recommended Protein Amount: ≥1 mg total protein (adjustable based on project needs)
3. Buffer Composition: Avoid high concentrations of detergents, salts, or alcohols
4. Storage and Shipping: Store at −80°C; ship on dry ice to maintain protein stability
5. Additional Information: Provide details such as sample source, treatment conditions, and research objectives for customized project design
Deliverables
1. Identification table of SUMOylated peptides and modification sites
2. Quality control report for enrichment and MS analysis
3. Functional annotation and pathway enrichment results
4. Raw MS data files
5. Comprehensive project report with visualized data outputs
SUMOylation site identification is a vital approach for deciphering the post-translational regulatory networks that control cellular processes. Leveraging advanced MS technologies, optimized enrichment strategies, and professional data analysis, MtoZ Biolabs delivers reliable and high-quality SUMOylation site identification service. Whether your research focuses on fundamental mechanisms or applied studies, we provide dependable technical support and actionable insights.
Contact MtoZ Biolabs today to learn how our SUMOylation site identification service can accelerate your proteomics research and uncover new dimensions of protein regulation.








