Disulfidomics Profiling Service
MtoZ Biolabs offers comprehensive Disulfidomics Profiling Services at the proteomics level, focusing on large-scale analysis of disulfide bonds in proteins. By leveraging advanced mass spectrometry (MS) and cutting-edge proteomics workflows, we provide a detailed, high-resolution map of disulfide connectivity across complex protein samples. Our service enables the identification, quantification, and structural characterization of disulfide bonds, supporting in-depth insights into protein structure, stability, and function at the omics level, essential for therapeutic protein development, disease research, and quality control in biopharmaceuticals.
What Is Disulfidomics?
Disulfidomics is a specialized branch of proteomics focused on the identification, characterization, and quantification of disulfide bonds within proteins. Disulfide bonds are covalent linkages formed between two cysteine residues, stabilizing the tertiary and quaternary structures of proteins by maintaining the correct conformation. These bonds are crucial for the structural integrity and function of many proteins, particularly those secreted into the extracellular environment, like antibodies, enzymes, and growth factors.
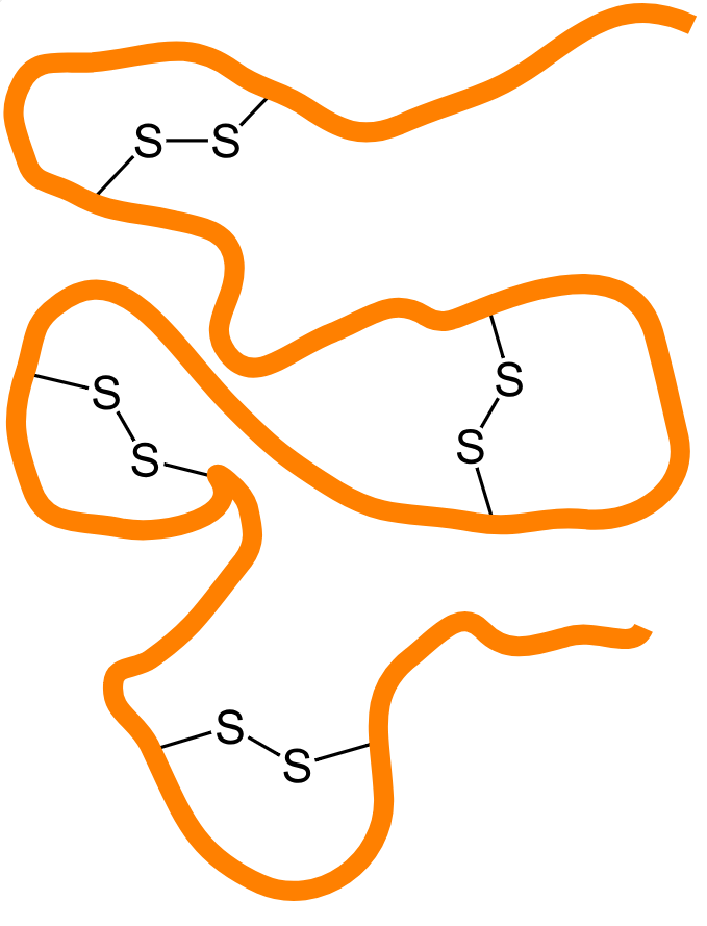
Sources: Wikipedia
Figure 1. Disulfide Bonds in Proteins
Workflow of Disulfidomics Profiling Service
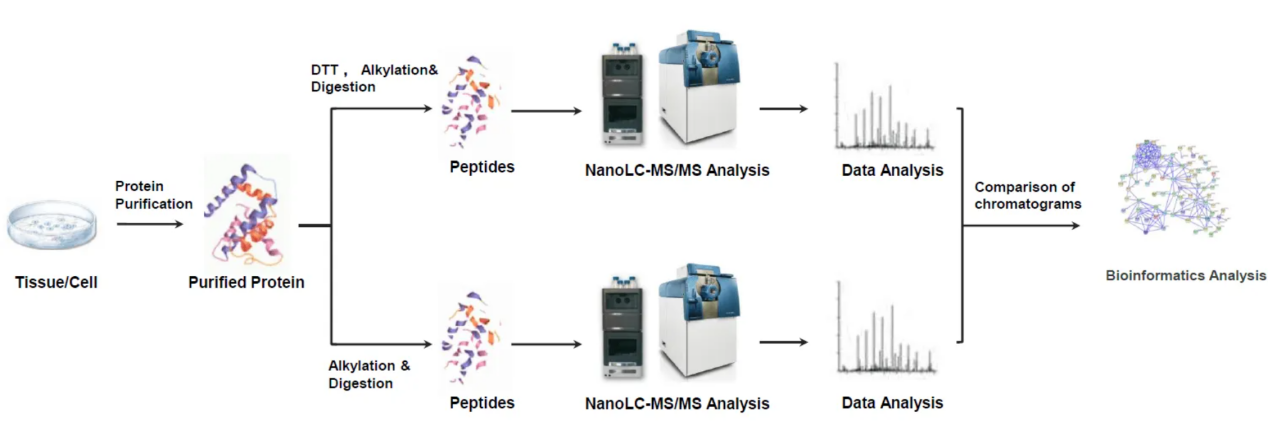
1. Protein Digestion and Sample Preparation
Proteins are typically denatured, reduced to break any existing disulfide bonds, and then digested with proteases such as trypsin to generate peptides for analysis. Following digestion, the sample undergoes a non-reducing alkylation step to prevent reformation of disulfide bonds, ensuring accurate detection.
2. Disulfide Bond Cleavage and Enrichment
After proteolytic digestion, the protein fragments are treated with specific disulfide bond-cleaving reagents such as DTT (dithiothreitol) or TCEP (tris(2-carboxyethyl)phosphine) to release the disulfide-linked peptides. These peptides are then enriched using affinity techniques to isolate only those containing disulfide linkages, making them detectable at high sensitivity levels.
3. LC-MS/MS Analysis
The enriched disulfide-containing peptides are separated using nano-liquid chromatography (nanoLC) and then analyzed with high-resolution mass spectrometry (MS/MS), typically using Orbitrap or Q-TOF systems. This allows for precise identification of peptide sequences and the disulfide bonds that crosslink them.
4. Data Processing and Structural Elucidation
Advanced software tools such as Proteome Discoverer, Byonic, and MaxQuant are used to process the mass spectrometry data. These tools allow for the identification of disulfide bond pairings, the elucidation of bond connectivity, and quantification of the abundance of disulfide-linked peptides.
5. Mapping and Validation
The results are verified through cross-validation with known disulfide bond patterns in existing databases or through manual inspection of the fragmentation spectra to ensure confidence in the disulfide connectivity assignment.
Why Choose MtoZ Biolabs
☑️Exceptional Sensitivity and Precision: By using high-resolution mass spectrometers, we provide highly accurate identification and quantification of disulfide bonds, ensuring reliable data even for low-abundance proteins.
☑️Comprehensive Disulfide Bond Profiling: Our service analyzes a wide range of proteins, capturing both abundant and low-abundance disulfide bonds to create an extensive connectivity map across your proteome of interest.
☑️Insight into Dynamic Disulfide Bond Changes: We enable monitoring of disulfide bond variations under different experimental conditions, offering critical insights into their regulatory role in protein function and stability.
☑️Advanced Data Interpretation: Through powerful bioinformatics platforms, we deliver in-depth analysis and interpretation of disulfide bond patterns, highlighting their biological significance and relevance to your research objectives.
Applications of Disulfidomics Profiling Service
1. Biopharmaceutical Development: Characterizing disulfide bonds in therapeutic proteins, antibodies, and enzyme preparations to ensure structural integrity and function.
2. Protein Engineering: Mapping disulfide bonds in engineered proteins to optimize folding, stability, and activity.
3. Biosimilar Comparability: Confirming identical disulfide bond patterns between biosimilars and reference products.
4. Disease Mechanism Research: Investigating misfolded proteins and disulfide bond disruptions in diseases such as Alzheimer's, Parkinson's, and cystic fibrosis.
5. Regulatory Compliance: Ensuring consistent disulfide bonding patterns across production batches for biopharmaceuticals to meet regulatory standards.
FAQ
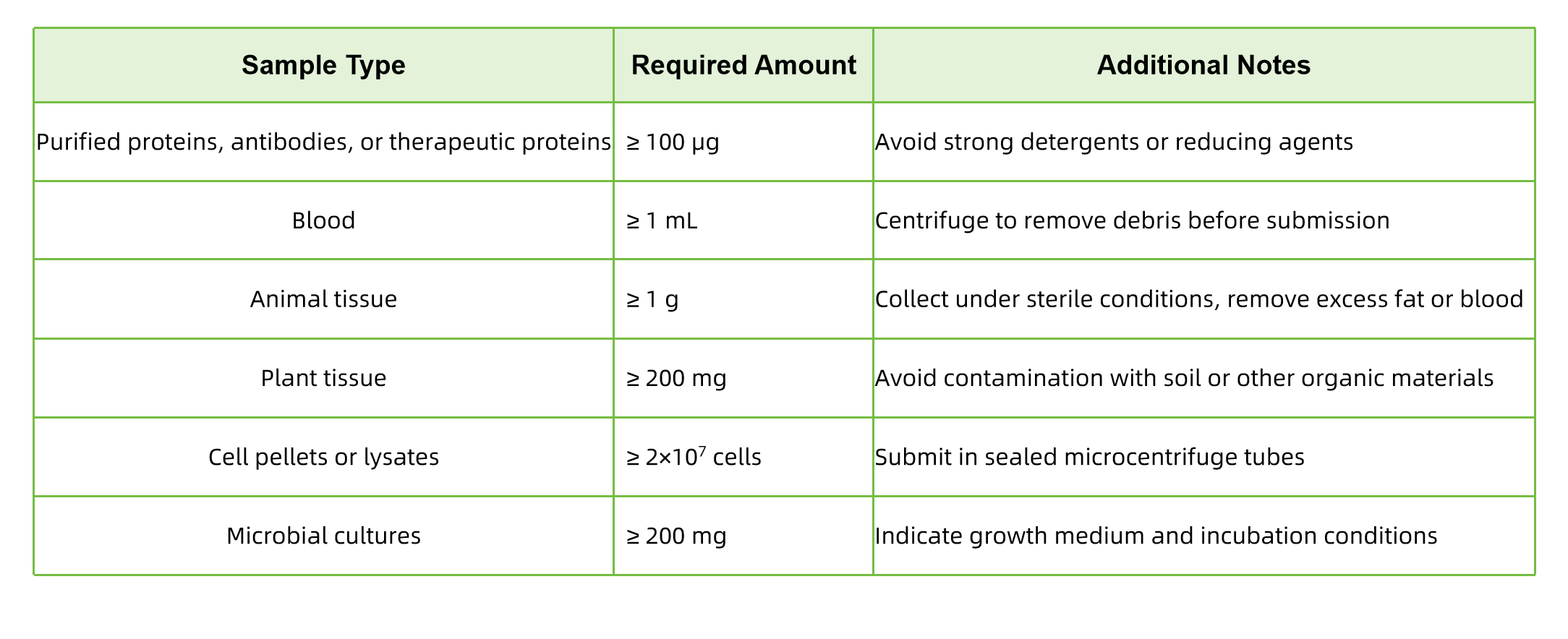
Q1: What types of samples are suitable?
MtoZ Biolabs accepts a wide range of biological samples, including but not limited to:
- Blood or other biological fluids
- Animal and plant tissues
- Cell pellets or lysates
- Microbial cultures
- Purified proteins
For other sample types, please contact us in advance for tailored preparation guidance.
Q2: How should I prepare my samples?
-
All samples should be stored at –80°C.
-
Use dry ice or ice packs during shipment to preserve protein integrity.
-
Avoid freeze-thaw cycles that may alter disulfide linkages.
For more information, please refer to Sample Submission Guidelines for Proteomics.
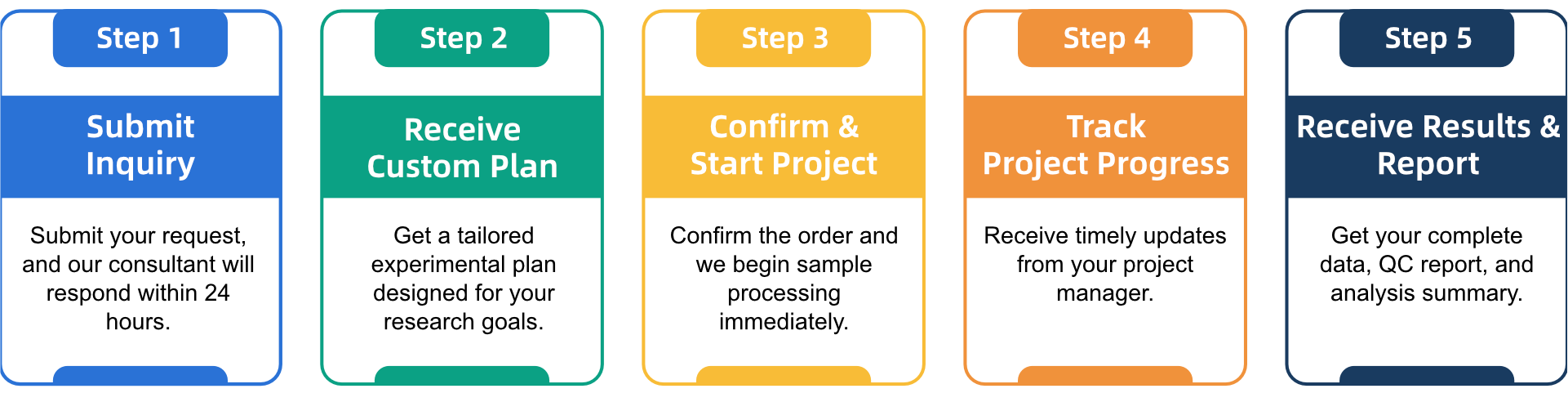
Q3: What is the service general workflow?
Q4: What data formats are provided?
Results are delivered in standard and easy-to-access formats, including:
-
Raw MS data files (.raw, .wiff, .d)
-
Processed data and quantification tables (.xlsx, .csv)
-
Spectral and chromatogram images (.pdf, .png)
-
Final analytical report (.pdf)
All deliverables are compatible with common data analysis software. Additional file formats or data structures can be provided upon request to meet specific research or publication requirements.
Start Your Project with MtoZ Biolabs
Contact us to discuss your experimental design or request a quote. Our technical specialists are available to provide a free business assessment.








