Protein Oxidation Analysis Service
Based on a high-resolution LC-MS/MS mass spectrometry and liquid chromatography separation platform, MtoZ Biolabs has launched the protein oxidation analysis service which can systematically detect and quantify oxidation modification sites in protein samples. This service, combined with specific enrichment and optimized experimental workflows, not only analyzes the types and distribution characteristics of oxidative modifications but also evaluates their abundance changes and potential functional impacts. The final output data include accurate identification results of oxidative modifications, quantitative difference information, and related functional annotations, providing reliable data support for researchers. MtoZ Biolabs provides analysis including but not limited to the following oxidation types:
✅ Protein Carbonylation
✅ S-glutathionylation
✅ Methionine Oxidation
✅ Dityrosine Crosslinking
✅ Tyrosine Nitration and S-nitrosylation
✅ Protein Radical Modifications
Overview
Protein oxidation refers to the process in which amino acid residues of proteins (such as cysteine, methionine, tryptophan, and tyrosine) undergo chemical modifications under the action of reactive oxygen species, free radicals, or metal ions. It is one of the common post-translational modifications occurring under cellular oxidative stress, aging, and environmental influences. This modification may alter the conformation, stability, and function of proteins, thereby affecting signal transduction, metabolic regulation, and cellular homeostasis. This method has been widely applied in antioxidant research, food and nutritional sciences, structural biology, and cellular function regulation, providing important data support for understanding the roles of proteins in environmental stress and physiological processes.
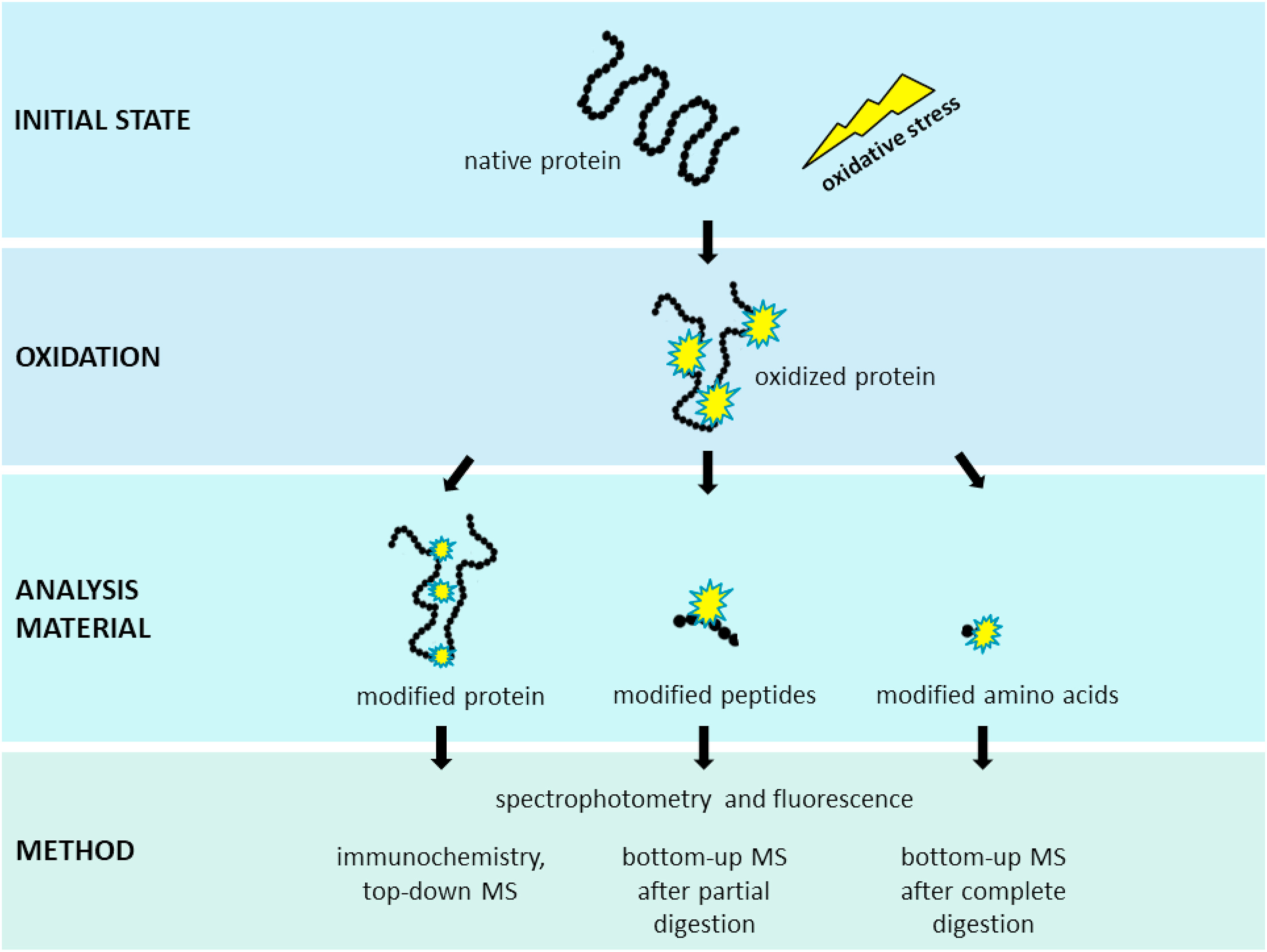
Kehm, R. et al. Redox Biology, 2021.
Figure 1. Analysis of Qxidative Protein Modifications.
Services at MtoZ Biolabs
1. Target Protein Oxidation Analysis
MtoZ Biolabs can perform site-level oxidation modification detection for designated target proteins, identifying different types of oxidative changes and comparing their differences under various conditions. Relying on high-resolution LC-MS/MS, we help researchers evaluate the specific impact of oxidation on protein structure and function.
2. Oxidation Proteomics Analysis
MtoZ Biolabs integrates enrichment strategies with high-throughput mass spectrometry to systematically identify and quantify oxidized peptides in complex samples. This service can reveal global oxidative regulatory features and support in-depth studies of stress responses, aging mechanisms, and disease-related oxidative changes.
Analysis Workflow
1. Sample Preparation
Proteins are extracted and quantified from cells, tissues, body fluids, or purified proteins to ensure uniform quality and provide a foundation for oxidative modification detection.
2. Protein Digestion
Proteins are digested with trypsin or other proteases into peptides to facilitate subsequent identification and localization of oxidative modifications.
3. Derivatization (if applicable)
For modifications such as protein carbonylation that are difficult to detect directly, chemical derivatization methods (such as DNPH labeling) are used to improve detection sensitivity and specificity.
4. Enrichment of Oxidized Peptides
Oxidized peptides are enriched through specific chemical capture, affinity chromatography, or selective labeling techniques, enhancing the signal-to-noise ratio and improving the detection of low-abundance modifications.
5. Mass Spectrometry Analysis
High-resolution LC-MS/MS combined with liquid chromatography is used to achieve precise identification and quantification of the enriched peptides, ensuring high sensitivity and confidence.
6. Data Analysis
Database searching and bioinformatics tools are applied to annotate oxidative modification sites, providing distribution patterns, abundance changes, and potential functional information to generate a comprehensive oxidation modification profile.
Service Advantages
1. High-Sensitivity Detection
Relying on a high-resolution LC-MS/MS platform, low-abundance oxidative modifications can be captured in complex backgrounds, ensuring reliable data.
2. Specific Enrichment
By combining derivatization and affinity chromatography, oxidized peptides can be effectively separated, improving the signal-to-noise ratio and detection accuracy.
3. Professional Team Support
Experiments and data analysis are performed by experts with extensive experience in proteomics and post-translational modification studies, ensuring standardized processes and reliable results.
4. Customized Solutions
The analysis depth and scope can be flexibly adjusted according to different research needs, providing targeted strategies for oxidative modification analysis.
Applications
1. Protein Stability Research
By detecting oxidation modification levels, the structural stability and degradation trends of proteins under different conditions can be evaluated.
2. Structural Biology Analysis
Protein oxidation analysis service can be used to reveal the effects of oxidative modifications on protein folding, conformation, and molecular interactions.
3. Cellular Function Regulation Research
Through the localization and quantification of oxidative modifications, their roles in signal transduction, metabolic processes, and stress responses can be analyzed.
4. Food and Nutritional Sciences
Protein oxidation analysis service can be applied to monitor protein oxidation changes during food processing or storage, assessing their impact on nutritional value and functionality.
FAQ
Q1: Does Protein Oxidation Require Derivatization Treatment?
A1: Not all modifications require it. Common modifications such as methionine oxidation and disulfide bond formation can be directly detected by LC-MS/MS; however, protein carbonylation often requires derivatization methods such as DNPH labeling due to its lower detection sensitivity.
Q2: Can Different Types of Oxidative Modifications Be Distinguished?
A2: Yes. By using high-resolution mass spectrometry combined with multiple fragmentation modes (such as HCD and ETD), various modifications including methionine oxidation, tyrosine nitration, and disulfide bond formation can be distinguished, and further confirmed through database annotation.
Q3: What Are the Requirements for Sample Storage and Transportation in Oxidative Modification Detection?
A3: Strict low-temperature storage (-80°C) is required, and repeated freeze-thaw cycles should be avoided. During transportation, dry ice or cold chain conditions must be used to ensure that modifications do not undergo additional changes due to environmental factors.








