4D-Ubiquitination Proteomics Service
Based on a high-resolution mass spectrometry platform combined with ion mobility separation technology, MtoZ Biolabs has launched the 4D-ubiquitination proteomics service to achieve systematic detection and quantitative analysis of ubiquitination modifications in complex biological samples. Through specific enrichment and optimized experimental workflows, this service can sensitively capture low-abundance ubiquitination sites and precisely analyze their distribution characteristics and abundance changes. The final output includes high-quality data such as modification site identification, quantitative differential analysis, and functional annotation, helping researchers comprehensively map ubiquitination modification profiles and providing reliable data support for cell signaling pathway analysis, drug target research, and biomarker discovery.
Overview
Ubiquitination is typically achieved by covalently attaching the small protein ubiquitin to the lysine residues of target proteins, thereby regulating their stability, degradation, localization, and interactions. It plays important roles in processes such as the cell cycle, signal transduction, DNA damage repair, and immune response. 4D-ubiquitination proteomics analysis enables high-sensitivity and systematic identification and quantification of ubiquitination sites in complex samples, generating comprehensive modification profiles that are widely applied in cell signaling pathway analysis, DNA damage repair mechanism studies, and the screening of drug targets and potential biomarkers.
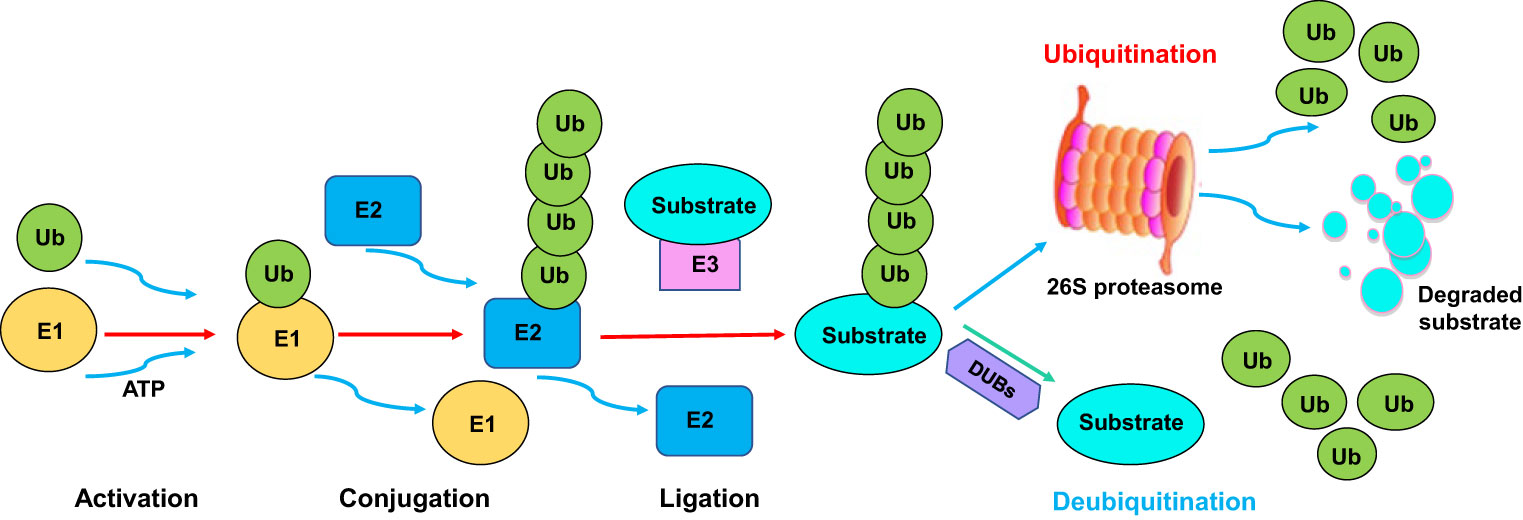
Zheng, C X. et al. Frontiers in Immunology, 2023.
Figure 1. The Ubiquitination and Deubiquitination Processes.
Analysis Workflow
1. Sample Preparation
Proteins are extracted and pretreated from experimental samples.
2. Protein Digestion and Phosphopeptide Enrichment
Proteins are digested under optimized conditions, and ubiquitinated peptides are selectively captured through specific enrichment strategies.
3. 4D Mass Spectrometry Detection
Relying on high-resolution mass spectrometry platforms, ubiquitinated peptides are identified and quantified with high sensitivity.
4. Data Analysis
By combining database searching with multidimensional bioinformatics analysis, outputs include ubiquitination site distribution, quantitative differences, and functional annotation, forming a systematic modification profile.
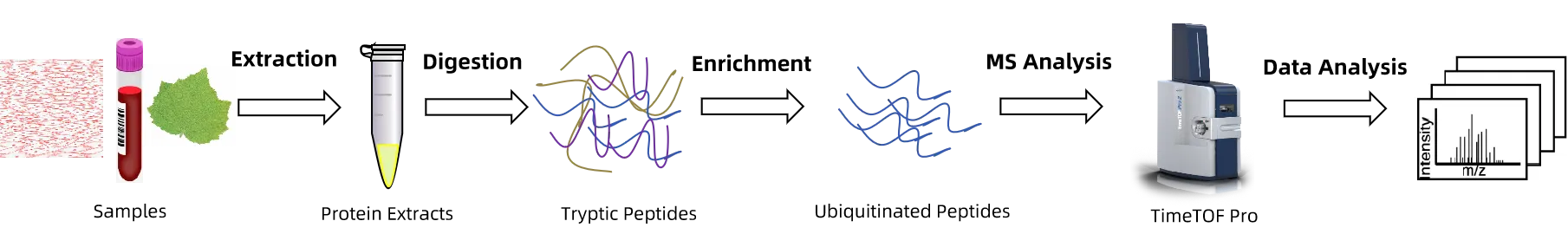
Figure 1. The Workflow of 4D-Ubiquitination Proteomics Analysis.
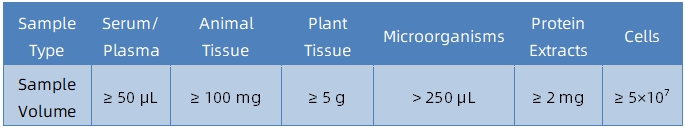
Sample Submission Suggestions
Service Advantages
1. High-Resolution Detection
Utilizing advanced LC-MS/MS combined with ion mobility separation technology, low-abundance ubiquitination sites are precisely captured to ensure data reliability.
2. Comprehensive Coverage
Systematic detection and quantification of multiple types of ubiquitination modifications are supported, providing a more complete modification profile.
3. High Sensitivity and Reproducibility
By integrating specific enrichment with optimized experimental workflows, detection sensitivity is enhanced, and data consistency and reproducibility are ensured.
4. Customized Solutions
Analysis strategies can be flexibly adjusted according to research objectives and sample characteristics to meet diverse application needs.
Applications
1. Cell Signaling Research
By analyzing ubiquitination modifications of key proteins, the regulatory roles in signal transduction can be revealed.
2. Protein Homeostasis and Degradation
The 4D-ubiquitination proteomics service can be used to evaluate the role of ubiquitination in maintaining protein dynamic balance and degradation pathways.
3. Structural and Functional Studies
Detection of ubiquitination sites supports the investigation of the relationship between protein structure and function.
4. Cell Cycle and Stress Response
The 4D-ubiquitination proteomics service can be applied to monitor changes in ubiquitination modifications at different stages or under stress conditions.
FAQ
Q1: What Is the Detection Depth of 4D Ubiquitination Proteomics?
A1: With optimized sample preparation and a high-sensitivity platform, routine experiments can identify thousands of ubiquitination sites. For complex tissues or enriched samples, 4D ion mobility separation can further expand coverage and capture low-abundance modifications.
Q2: Can Ubiquitination Modifications Be Precisely Defined at the Site Level?
A2: Yes. By combining specific enrichment (such as K-ε-GG residue labeling) with multidimensional separation, precise localization of ubiquitination sites can be achieved, along with quantitative information at the residue level.








