Histone Succinylation Analysis Service
Based on a high-resolution liquid chromatography–tandem mass spectrometry (LC-MS/MS) platform, the histone succinylation analysis service launched by MtoZ Biolabs enables systematic detection and quantitative analysis of succinylation modifications on histone lysine residues. Through optimized sample preparation and specific enrichment strategies, this service achieves precise identification and abundance analysis of low-level modification sites. By integrating bioinformatics tools, it provides high-quality data on modification site distribution, differential comparison, and functional annotation, helping researchers elucidate the role of succinylation in epigenetic regulation and metabolic modulation.
Overview
Histone succinylation (Ksucc) refers to a post-translational modification in which a succinyl group is covalently attached to the lysine residues of histones. This modification significantly alters the charge and conformation of proteins, thereby influencing chromatin structure and gene transcription activity. Ksucc is closely associated with cellular metabolic processes, particularly energy homeostasis and the tricarboxylic acid (TCA) cycle. Histone succinylation analysis enables the identification and quantification of modification sites, revealing their roles in epigenetic regulation, metabolic control, and cellular function maintenance, and providing strong support for studying the link between metabolism and epigenetic regulation.
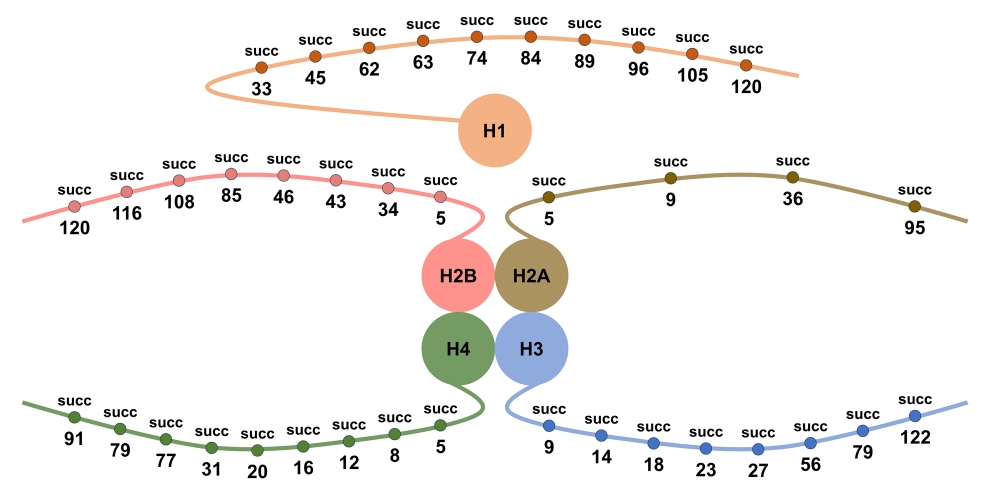
Zhang, M H. et al. Genome Instability & Disease, 2025.
Figure 1. Known Histone Succinylation Sites Reported in Existing Literature.
Analysis Workflow
1. Histone Extraction and Digestion
Histones are extracted from samples and digested under optimized conditions to generate analyzable peptides.
2. Modified Peptide Enrichment
Specific enrichment methods are used to selectively capture succinylated peptides.
3. LC-MS/MS Analysis
High-resolution mass spectrometry is employed to accurately identify and quantify modification sites.
4. Data Analysis
Bioinformatics tools are applied to analyze modification distribution and abundance changes, generating comprehensive research reports.
Sample Submission Suggestions
1. Sample Type and Quantity
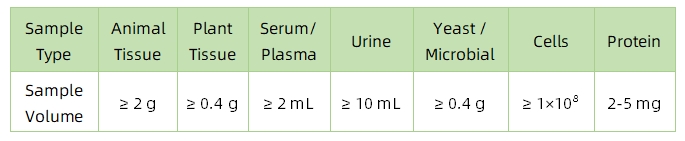
Note: Plasma should be collected using EDTA as an anticoagulant. Standard tissue or cell lysis buffers can be used during protein extraction.
2. Sample Transportation
Avoid repeated freeze-thaw cycles. Samples are recommended to be stored at -80°C and transported on dry ice to ensure low-temperature conditions throughout the process and prevent modification loss.
Note: For special samples or if a detailed submission plan is required, please contact MtoZ Biolabs technical staff in advance.
Service Advantages
1. High-Resolution Mass Spectrometry Analysis
Employ advanced LC-MS/MS technology to ensure high-precision identification and quantification of succinylation modifications.
2. Comprehensive Site Coverage
Achieve systematic detection and analysis of multiple histone subtypes and succinylation sites.
3. Expert Technical Team
Technical support and data interpretation are provided by experienced post-translational modification (PTM) proteomics experts.
4. Flexible Customized Solutions
Design personalized solutions according to sample type and research objectives to meet diverse experimental needs.
Applications
1. Epigenetic Regulation Research
The histone succinylation analysis service can be used to investigate the role of succinylation in chromatin remodeling and transcriptional regulation.
2. Cell Development Research
By analyzing succinylation during cell differentiation and development, researchers can explore its dynamic regulation.
3. Signal Transduction and Stress Response Research
The histone succinylation analysis service can help study how succinylation participates in regulatory mechanisms under environmental stimuli or oxidative stress.
4. Biomarker Screening Research
Analyzing succinylation under specific physiological or metabolic states can assist in identifying potential molecular biomarkers.
FAQ
Q1: What Sites Can Be Detected in Histone Succinylation Analysis?
A1: This service covers succinylation modification sites on multiple histone subtypes (such as H3 and H4), including common sites like H3K79succ and H4K12succ, and can be expanded to additional targets as required by the research.
Q2: What Are the Differences between Succinylation, Acetylation, and Malonylation?
A2: Succinylation introduces a larger substituent group carrying a negative charge, which significantly alters the protein's charge and spatial conformation. Compared with acetylation and malonylation, it exerts a stronger impact on chromatin structure and protein function.
Q3: Can Multiple Post-Translational Modifications Be Analyzed Together?
A3: Yes. Succinylation can be studied in combination with acetylation, methylation, propionylation, and other modifications to reveal potential synergistic or antagonistic interactions between different modification types.








