Asymmetric Dimethylation Analysis Service
MtoZ Biolabs provides Asymmetric Dimethylation Analysis Service to support researchers in accurately identifying, localizing, and quantifying asymmetric dimethylation (aDMA) modifications on proteins. Utilizing advanced Orbitrap Fusion Lumos and Q Exactive HF mass spectrometers, combined with optimized enrichment techniques and tailored bioinformatics pipelines, this service delivers high-resolution data for studying the role of asymmetric dimethylation in gene regulation, protein function, and disease mechanisms. By overcoming the analytical challenges associated with low-abundance modifications, MtoZ Biolabs provides reproducible and publication-ready datasets that drive both fundamental and translational research.
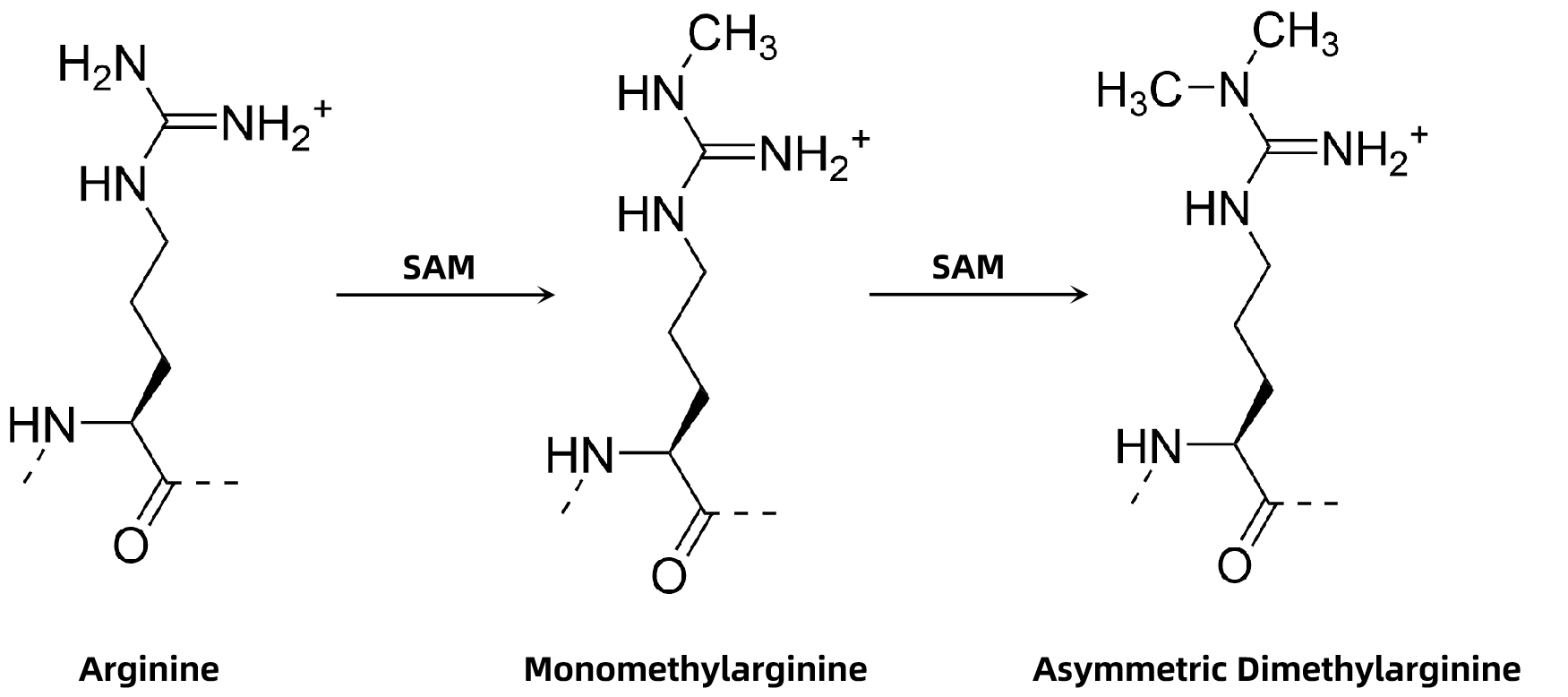
Figure 1. Arginine Asymmetric Dimethylation
What is Asymmetric Dimethylation?
Asymmetric dimethylation is a type of arginine methylation in which two methyl groups are added to one of the terminal nitrogen atoms of the guanidino group. This modification is catalyzed by type I protein arginine methyltransferases (PRMTs) and is distinct from symmetric dimethylation (sDMA). aDMA plays critical roles in regulating RNA processing, signal transduction, transcriptional control, and protein-protein interactions. Aberrant aDMA has been linked to cancer progression, autoimmune diseases, and neurological disorders, making its precise characterization vital for biomedical research and therapeutic development.
Services at MtoZ Biolabs
1. Targeted Protein Asymmetric Dimethylation Analysis
MtoZ Biolabs offers targeted analysis of asymmetric dimethylation on client-specified proteins, providing precise site-level identification and quantitative evaluation of defined targets. This service is suited for studies focusing on specific regulatory proteins, targeted validation, or assessment of modification changes under controlled experimental conditions.
2. Asymmetric Dimethylation Proteomics
For broader applications, MtoZ Biolabs provides asymmetric dimethylation proteomics to profile dimethylation events across complex biological samples. This global approach supports discovery-driven research, comparative studies across treatment groups or biological states, and the identification of asymmetric dimethylation patterns with potential functional or biomarker relevance.
Analysis Workflow
The Asymmetric Dimethylation Analysis Service at MtoZ Biolabs is carried out through a carefully optimized process:
1. Sample Preparation: Proteins are extracted under strict conditions to maintain modification stability.
2. Enzymatic Digestion: Proteins are digested with trypsin or other proteases to generate peptides suitable for downstream analysis.
3. Targeted Enrichment: Immunoaffinity purification or chemical enrichment is applied to selectively isolate asymmetric dimethylated peptides.
4. LC-MS/MS Detection: Ultra-high-resolution Orbitrap mass spectrometry ensures accurate identification and quantification of modification sites.
5. Computational Analysis: Bioinformatics pipelines map modification sites, quantify abundance, and integrate pathway-level information.
6. Quality Validation: Internal standards and replicate analysis confirm data reliability and reproducibility.
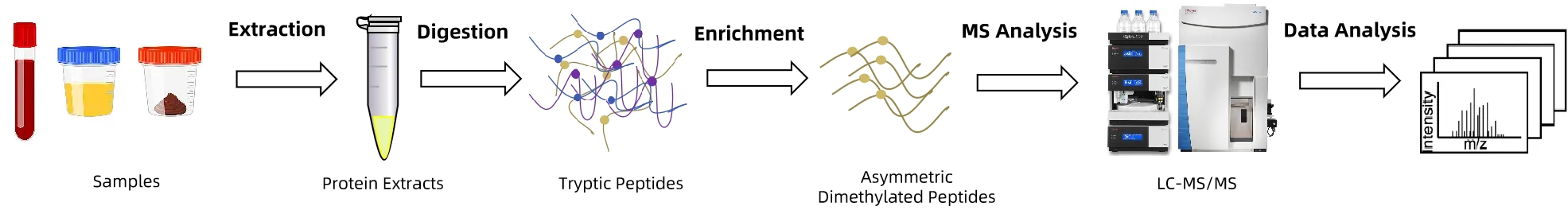
Figure 2. Asymmetric Dimethylation Analysis Workflow
Sample Submission Suggestions
MtoZ Biolabs accepts multiple sample types for asymmetric dimethylation analysis:
Serum/Plasma: ≥100 µL
Urine: ≥2 mL
Animal Tissue: ≥200 mg
Plant Tissue: ≥10 g
Microorganisms: ≥500 µL
Protein Extracts: ≥5 mg
Cells: ≥1 × 108
Samples should be frozen at -80°C and shipped on dry ice. Detergents, salts, or stabilizers that interfere with MS analysis should be avoided. If you need further details, our technical support team is happy to assist and provide comprehensive guidance on sample submission.
Applications
The Asymmetric Dimethylation Analysis Service is widely applicable across research fields:
Epigenetics: Profiling arginine methylation in histones to explore chromatin regulation and transcriptional activity.
Cancer Research: Investigating the role of abnormal aDMA in tumor initiation, progression, and therapy resistance.
Neurobiology: Studying how asymmetric dimethylation influences neuronal gene regulation and neurodegenerative diseases.
Autoimmune Disorders: Characterizing aDMA-associated signaling pathways involved in immune regulation.
Drug Discovery: Identifying potential therapeutic targets by mapping aDMA across disease-relevant proteins.
Why Choose MtoZ Biolabs?
✅ Advanced PTM Analysis Platform: Integrated mass spectrometry systems and optimized enrichment ensure high-confidence identification of dimethylation sites.
✅ High Data Quality: Strict QC standards and in-depth bioinformatics interpretation ensure robust and reproducible datasets.
✅ Broad Sample Compatibility: Workflows validated for cells, tissues, bodily fluids, and protein extracts.
✅ Customized Solutions: Tailored experimental design and flexible data analysis to meet diverse research objectives.
✅ Transparent Pricing: One-time charge model with no hidden fees, providing cost clarity for research planning.
What Could be Included in the Report?
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
FAQ
Q1: What methods are used to differentiate asymmetric from symmetric dimethylation?
Asymmetric and symmetric dimethylation yield distinct ion fragmentation patterns. At MtoZ Biolabs, high-resolution Orbitrap MS combined with optimized dissociation methods ensures confident discrimination between these two modifications.








