Histone Methionylation Analysis Service
Based on a high-resolution liquid chromatography-tandem mass spectrometry (LC-MS/MS) platform, the histone methionylation analysis service launched by MtoZ Biolabs enables precise detection and quantitative analysis of methionylation modifications in histones. Through optimized sample preparation and highly sensitive mass spectrometric detection, the service identifies modification sites and evaluates abundance variations. Combined with bioinformatics analysis, it provides data on modification site distribution, quantitative differences, and functional annotations, supporting research on the roles of methionylation in epigenetic regulation and metabolic homeostasis.
Overview
Histone methionylation is a newly identified post-translational modification (PTM) in which a methionyl or methionine-related structural group is covalently attached to specific sites on histones, potentially influencing chromatin conformation and gene transcriptional activity. This modification is believed to be closely associated with cellular redox states, metabolic regulation, and protein conformational stability, and has been widely applied in studies of epigenetics, metabolic adaptation, and cellular stress responses.
Analysis Workflow
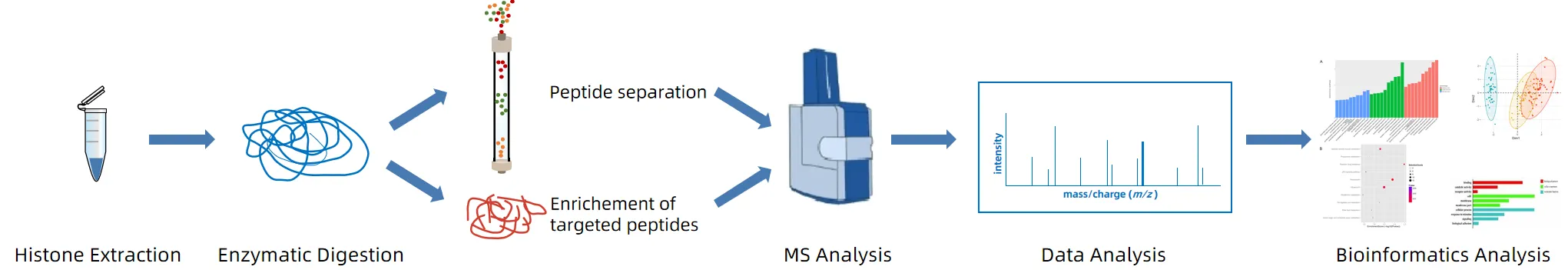
Figure 1. The Workflow of Histone Methionylation Analysis.
Sample Submission Suggestions
1. Sample Type and Quantity
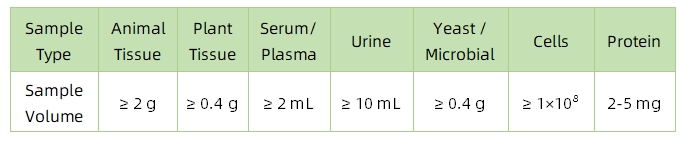
Note: Plasma should be collected using EDTA as an anticoagulant. Standard tissue or cell lysis buffers can be used during protein extraction.
2. Sample Transportation
Avoid repeated freeze-thaw cycles. Samples are recommended to be stored at -80°C and transported on dry ice to ensure low-temperature conditions throughout the process and prevent modification loss.
Service Advantages
1. High-Resolution Detection
Utilizing an advanced LC-MS/MS platform, the service precisely identifies low-abundance methionylation modifications with high sensitivity for accurate quantification.
2. Efficient Sample Processing
Employing optimized histone extraction and enzymatic digestion workflows ensures the integrity and stability of methionylation modifications throughout analysis.
3. Rigorous Quality Control
Standardized procedures and multi-level quality control are implemented across all steps to guarantee data accuracy and reproducibility.
4. Customized Service Solutions
Experimental workflows are flexibly designed according to sample type and research objectives to accommodate diverse analytical needs.
Applications
1. Epigenetic Regulation Studies
Histone methionylation analysis service can be used to investigate the role of methionylation in chromatin structural regulation and transcriptional activity control.
2. Cellular Metabolism Research
By studying methionylation events within cellular metabolism, researchers can explore its associations with redox balance and energy homeostasis.
3. Protein Function Regulation
Histone methionylation analysis service enables the study of how methionylation affects protein folding, conformational stability, and molecular interactions.
4. Signal Transduction Analysis
By monitoring modification dynamics, the service helps reveal regulatory mechanisms of methionylation in signaling pathways and stress responses.
FAQ
Q1: Is Methionylation Prone to Oxidation during Sample Preparation?
A1: Methionine residues are indeed susceptible to oxidation during processing. To prevent artificial modifications, antioxidants, low-temperature conditions, and inert atmospheres are applied, followed by reductive recovery and stability verification steps to ensure data authenticity.
Q2: Can This Service Be Integrated with Other Epigenetic Modification Analyses?
A2: Yes. It can be combined with analyses of methylation, acetylation, oxidation, and other post-translational modifications to elucidate complex regulatory networks.
Q3: Is Cross-Omics Integration Supported?
A3: Supported. The service allows integration with transcriptomic, metabolomic, and protein–protein interaction datasets to explore the systemic regulatory functions of methionylation.








